Molecular typing method for distinguishing different strains of cronobacter through performing single enzyme digestion on gluA gene based on RsaI
A Cronobacter, molecular typing technology, applied in microorganism-based methods, biochemical equipment and methods, microbial determination/inspection, etc., can solve the problem of inability to effectively distinguish different Cronobacter Rapid identification of the source of bacterial contamination and high technical operation requirements can achieve the effect of clear enzyme cut bands, stable identification and high technical requirements.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
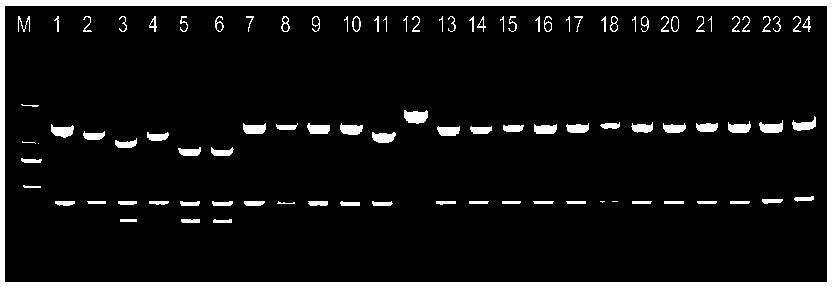
[0023] A single enzyme digestion based on RsaI gluA The molecular typing method for genetically differentiating the different species of Cronobacter spp. is a two-stage experimental phase, the first as figure 1 Shown, amplified Cronobacter specific gluA Gene fragments, the second stage as figure 2 As shown, the Cronobacter specific gluA Gene fragments were obtained to obtain different enzyme digestion maps, and then to distinguish six different species of Cronobacter genus.
[0024] The specific operation steps are as follows:
[0025] (1) selection gluA Gene
[0026] gluA The gene encodes Cronobacter α-glucosidase.
[0027] (2 pairs gluA gene amplification
[0028] Use amplification primers F: 5'- AGGGATCCTGAAAGCAATCGACAAGAA -3'; R: 5'-CCGAAGCTTACTCATTACCCTCCTGATG -3' pair gluA Gene amplification to obtain six different bacteria of the genus Crobacter gluA Gene fragments, see figure 1 : Strains of six different species of Cronobacter gluA Amplified re...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com