Preparation and application of ptc stable cell line using optimized gene codon extension system
A codon and translation system technology, applied in the field of biopharmaceuticals, can solve problems such as high cost, cumbersome construction process, and low virus rescue efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0127] Example 1: Construction and acquisition of Tol2-RS-IRES-*GFP-12tRNA-puro transposable vector
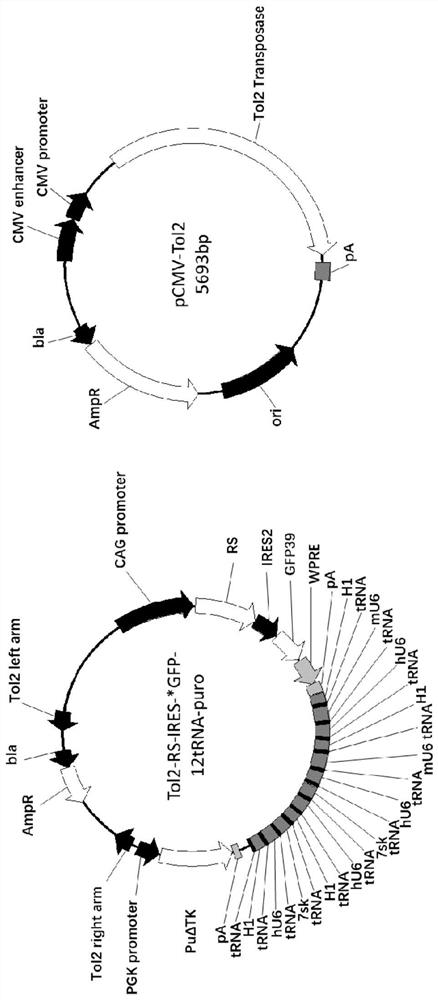
[0128] In order to ensure the expression of bio-orthogonal tRNA and aminoacyl-tRNA synthetase, the screening efficiency of positive cells and the integration efficiency of genes, it is necessary to combine multiple copies of promoter-tRNA, pylRS and single-point mutation GFP reporter gene in tandem Cloned into a transposable vector.
[0129] Therefore, the inventor has designed such as figure 1 In the Tol2-RS-IRES-*GFP-12tRNA-puro transposon vector shown in the middle left panel, the aminoacyl tRNA synthetase initiated by the CAG promoter was first introduced into the Tol2-puro transposon vector, and the internal ribosome entry site was used to The mutant GFP reporter gene with premature stop codon was introduced by dot connection; at the same time, 12 copies of the tRNA promoted by the PolIII promoter were designed, and the multiple copies of the tRNA fragments in series ...
Embodiment 2
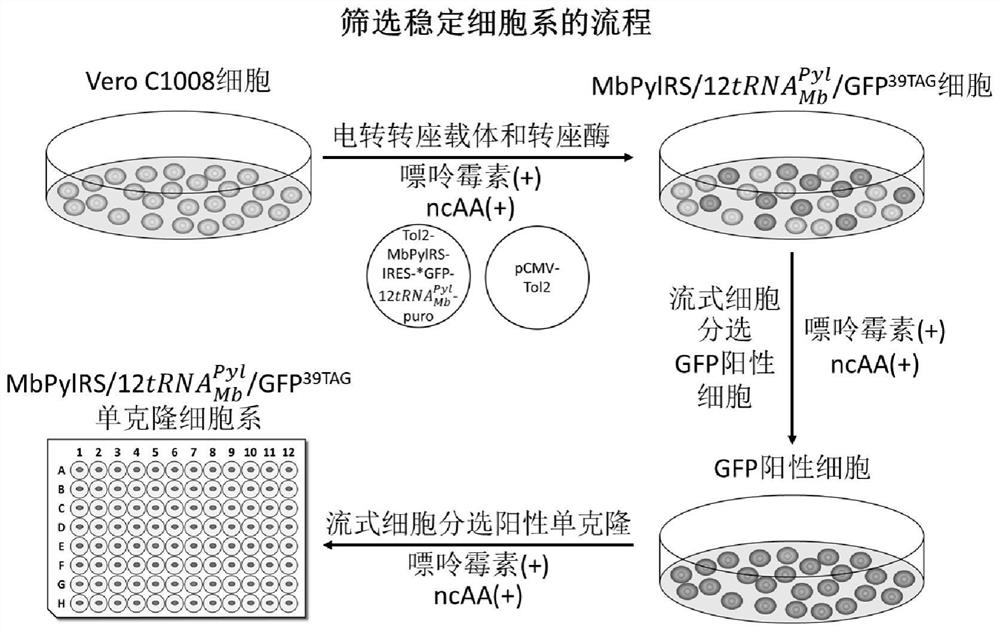
[0130] Example 2: Screening of Vero-Tol2-NAEK stable cell lines
[0131] (1) Preparation of cell suspension:
[0132] Vero cells (ATCC, CCL-81) were cultivated to a confluence of 70 with complete medium (MEM, Gibco, 11095080; 10% fetal bovine serum, PAN, P30-3302; 1% penicillin / streptomycin, Macgene, CC004). When % to 80%, the cells were digested and collected, rinsed three times with Opti-MEM (Gibco, 31985070) to wash away the antibiotics and serum in the medium, then resuspended with Opti-MEM, and the cells were blown up and down to make them free of clumping. Take a small amount of suspension for cell count and calculate the total cell number.
[0133] (2) Prepare the mixture of cells and plasmids:
[0134] Mix a certain amount of cells with plasmid DNA, and mix thoroughly to make the final concentration reach 1×10 in each tube of 100 μl mixture. 6 Cells and 10 μg plasmid DNA (the ratio of transposable vector to transposase plasmid DNA is 3:2), in which the cell volume...
Embodiment 3
[0143] Embodiment 3: Identification of Vero-Tol2-ncAA stable cell line (taking Vero-Tol2-NAEK as an example)
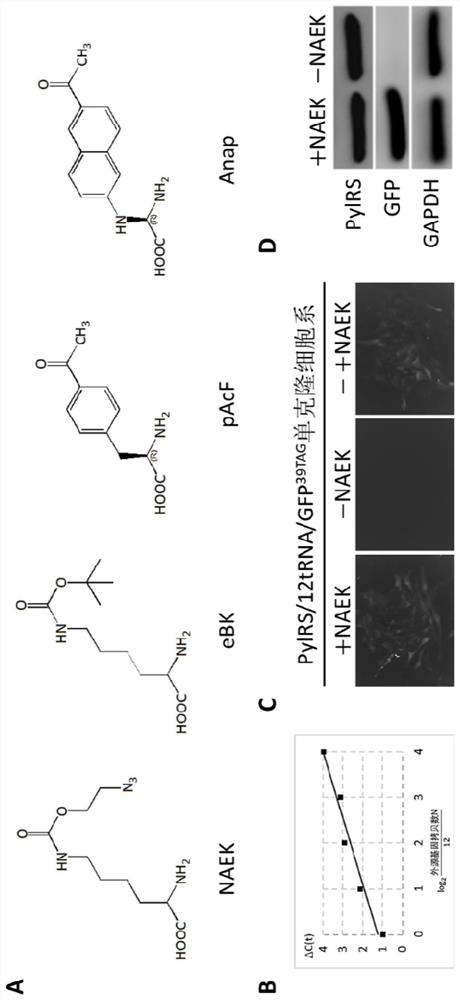
[0144] The stable cell line Vero-Tol2-NAEK constructed in the present invention contains tRNA derived from Methanosarcina pasteurii and pyrrolysyl-tRNA synthetase (MbPylRS), in protein expression in stable cell lines, with premature stop codons (including TAG, TAA, and TGA) as sense codons, capable of incorporation of the unnatural amino acid NAEK into protein. Next, the inventors examined the incorporation possibility of NAEK and the production performance of the mutant protein.
[0145] (1) Synthesis and identification of unnatural amino acid NAEK:
[0146] The chemical synthesis reaction formula of unnatural amino acid NAEK is as follows:
[0147]
[0148] As described in the above formula, 2.3 mL of raw material 1 (2-bromoethanol) was dissolved in a mixed solution of 90 mL of acetone and 15 mL of water, 3.12 g of NaN3 was added, and the mixture was heated ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com