Application and correlation product of human IMPA2 gene
A gene and application technology, applied in the field of application of human IMPA2 gene and related products, can solve problems such as the absence of IMPA2 gene
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0102] Example 1 Preparation of RNAi lentivirus for human IMPA2 gene
[0103] 1. Screening for effective siRNA targets against the human IMPA2 gene
[0104] Retrieve IMPA2 (NM_014214) gene information from Genbank; design effective siRNA targets for IMPA2 gene. Table 1-1 lists the screened effective siRNA target sequences against the IMPA2 gene.
[0105] Table 1-1 siRNA target sequence targeting human IMPA2 gene
[0106] SEQ ID NO TargetSeq(5'-3') 1 CTTACAGACGATTAACTAT
[0107] 2. Preparation of lentiviral vector
[0108] Aim at the siRNA target (take SEQ ID NO: 1 as an example) to synthesize a double-stranded DNA Oligo sequence (Table 1-2) with Age I and EcoR I restriction sites at both ends; Dicer acts on the pGCSIL-GFP vector (provided by Shanghai Jikai Gene Chemical Technology Co., Ltd.) to linearize it, and agarose gel electrophoresis identifies the digested fragment.
[0109] Table 1-2 Double-stranded DNA Oligo with sticky ends containing Age I...
Embodiment 2
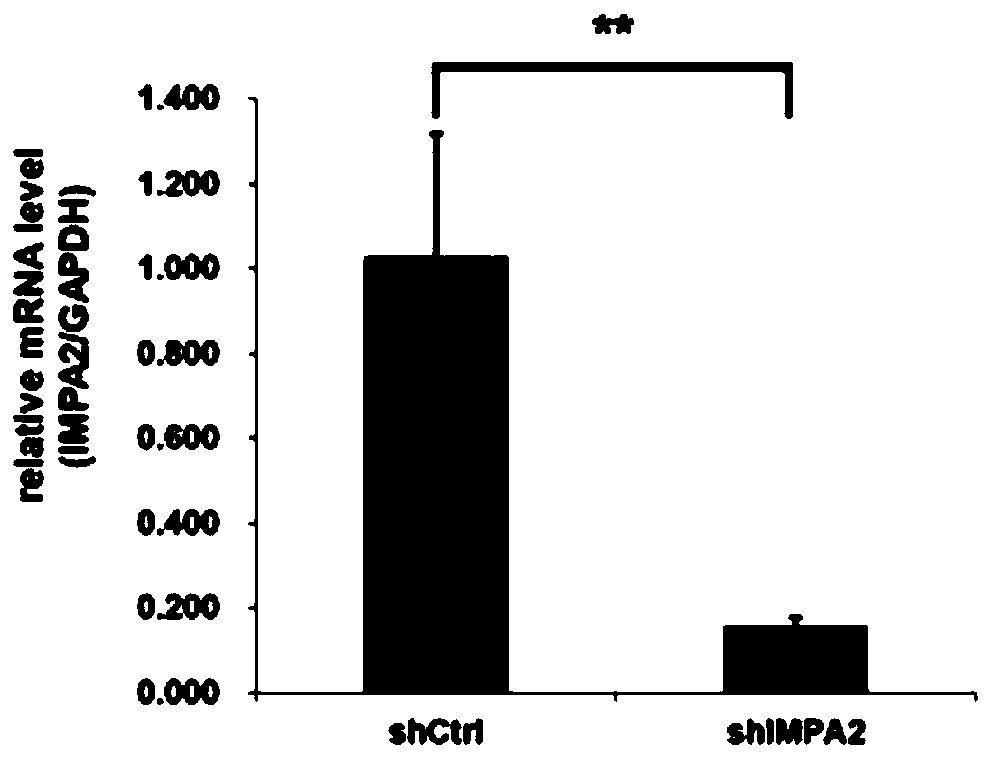
[0128] Example 2 Detection of silencing efficiency of genes infected with IMPA2-siRNA lentivirus
[0129] Human cervical cancer SiHa cells in logarithmic growth phase were trypsinized to make cell suspension (the number of cells was about 5×10 4 / ml) were inoculated in a 6-well plate and cultured until the cell confluency reached about 30%. According to the multiplicity of infection (MOI, SiHa: 20), an appropriate amount of the lentivirus prepared in Example 1 was added, the culture medium was replaced after 24 hours of culture, and the cells were collected after the infection time reached 5 days.
[0130] a) Detection of gene silencing efficiency by real-time fluorescent quantitative RT-PCR
[0131] Total RNA was extracted according to the instruction manual of Invitrogen's Trizol. According to the M-MLV instruction manual of Promega Company, RNA was reverse-transcribed to obtain cDNA (see Table 2-1 for the reverse transcription reaction system, react at 42°C for 1 hour, ...
Embodiment 3
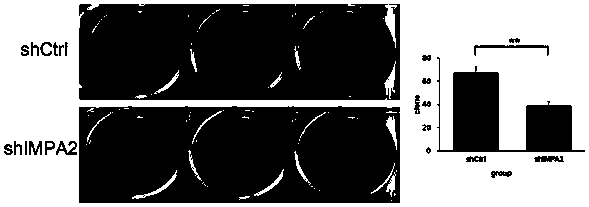
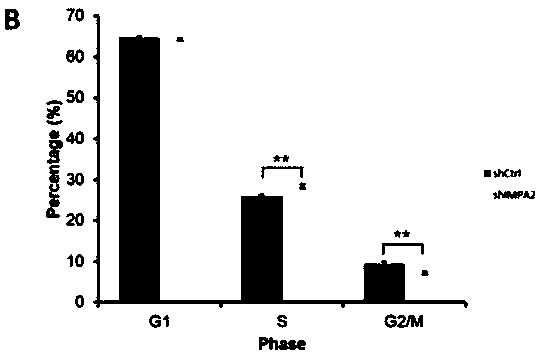
[0168] Example 3 Detection of proliferation ability of tumor cells infected with IMPA2-siRNA lentivirus
[0169] Human cervical cancer SiHa cells in logarithmic growth phase were trypsinized to make cell suspension (the number of cells was about 5×10 4 / ml) were inoculated in a 6-well plate and cultured until the cell confluency reached about 30%. According to the multiplicity of infection (MOI, SiHa: 20), an appropriate amount of virus was added, and the culture medium was replaced after 24 hours of culture. After the infection time reached 5 days, the cells of each experimental group in the logarithmic growth phase were collected. The complete medium was resuspended into a cell suspension (2×10 4 / ml), inoculate a 96-well plate at a cell density of about 1000 / well. 5 replicate wells in each group, 100 μl per well. After laying the board, place at 37°C, 5% CO 2Incubator cultivation. From the second day after plating, the plate was detected and read once a day with a Celi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com