Novel specific molecular targets for 16 different serotypes of salmonella and rapid detection method thereof
A technology for Salmonella and Salmonella enteritidis, which is applied in the field of microbiological testing, can solve the problems of no PCR method establishment report, and the report of PCR detection method for new molecular targets specific to serotype Salmonella has not been found, and achieves short detection time, simple result judgment, low cost effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0062] Example 1 Mining of new molecular targets specific to different species of Salmonella
[0063] According to the GenBank database and the complete genome DNA sequences of different serotypes of Salmonella self-tested by our team, bioinformatics analysis was carried out; 16 different serotypes of Salmonella (Salmonella Enteritidis, Salmonella Typhimurium, Salmonella Delphi, Salmonella Indiana, Argonne Salmonella, Salmonella infantis, Salmonella lyssen, Salmonella Brundenloop, Salmonella Vershaw, Salmonella Albany, Salmonella Hadar, Salmonella Wertfrieden, Salmonella London, Salmonella turkey, Salmonella Senftenberg, and Salmonella Pomona ) specific gene fragment, the nucleotide sequence of said gene fragment is shown in SEQ ID NO.1-SEQ ID NO.72.
[0064] Wherein, the sequences SEQ ID NO.1-4 are used to identify the Salmonella enteritidis, the sequence SEQ ID NO.5 is used to identify the Salmonella typhimurium, and the sequences SEQ ID NO.6-13 are used to identify the Salm...
Embodiment 2
[0065] Example 2 Rapid Detection of Primer Effectiveness
[0066] 1) Primer design
[0067] According to the sequence SEQ ID NO.1-72 in Example 1, a specific PCR amplification primer set (including a forward primer and a reverse primer) was designed. The sequence of the primer set is shown in Table 1.
[0068] Table 1 Specific PCR detection primer set
[0069]
[0070]
[0071]
[0072]
[0073]
[0074]
[0075]2) the method for identifying said 16 different serotypes of Salmonella, the steps are as follows:
[0076] S1 DNA template preparation: Use a commercial bacterial genomic DNA extraction kit to extract the bacterial genomic DNA to be tested as the template DNA to be tested;
[0077] S2 PCR amplification: use one of the primer sets 1-72 to perform PCR amplification on the DNA of the sample to be tested;
[0078] ①PCR detection system:
[0079]
[0080]
[0081] in:
[0082] The primer sets 1-4 are used to identify the Salmonella enteritidis,...
Embodiment 3
[0095] The specificity evaluation of embodiment 3 Salmonella enteritidis PCR detection method
[0096] Take 91 strains of Salmonella enteritidis, 41 strains of Salmonella of non-target serotypes, and 22 strains of non-Salmonella, and perform PCR detection according to the method in Example 2. Wherein, the S1 DNA template is prepared by extracting the genomic DNA of each bacterium; during the S2PCR amplification, the primers used are any one of the primer sets 1-4.
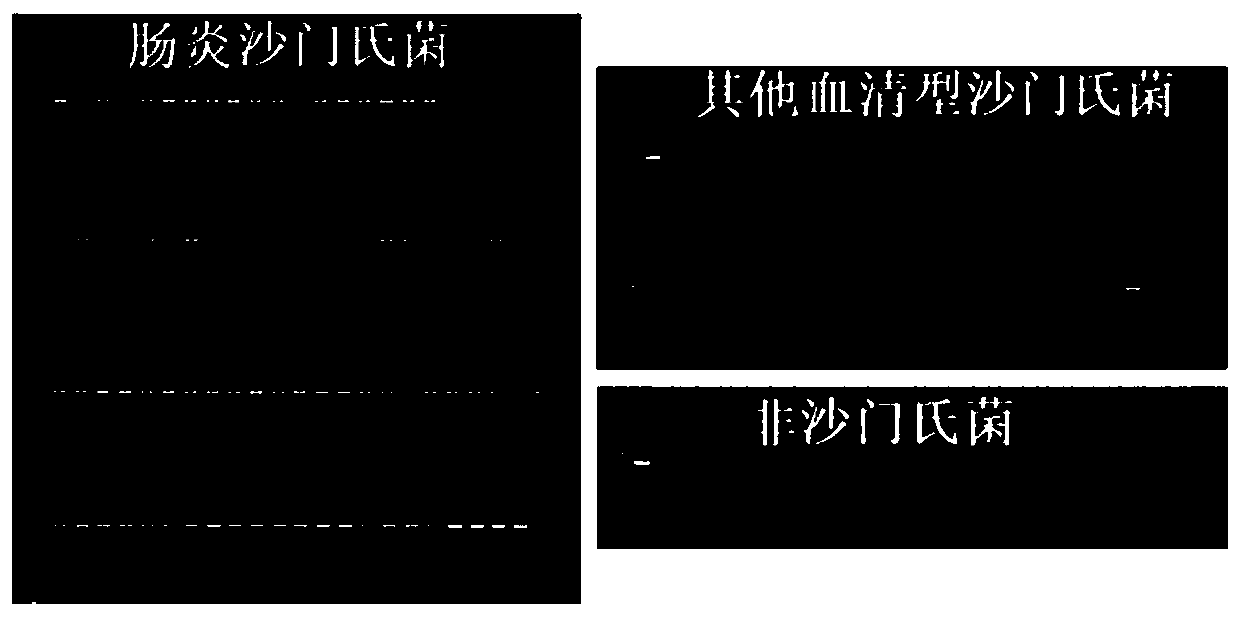
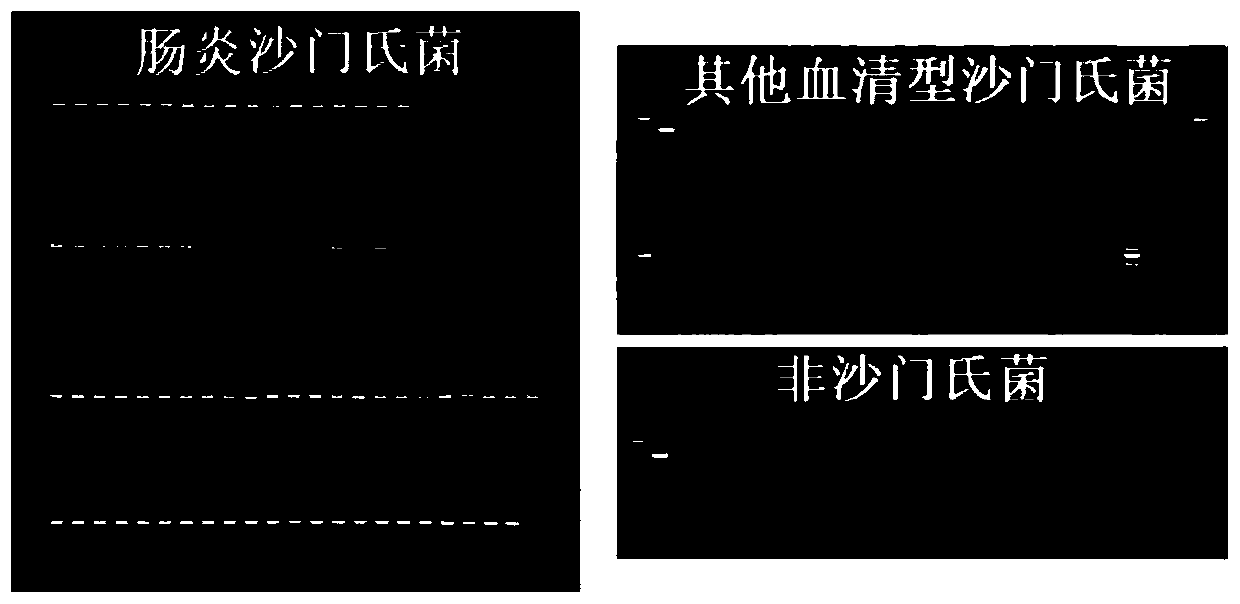
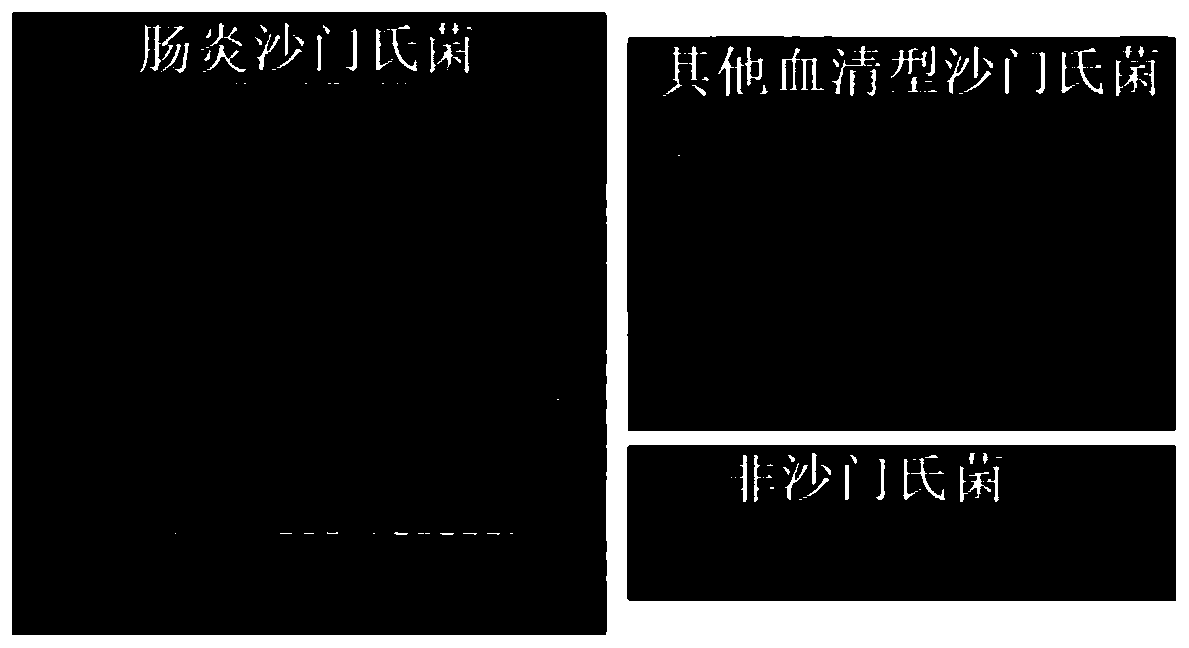
[0097] The gel results of PCR amplification products are as follows: Figure 1~4 (The first and last ones in the gel graph of Salmonella Enteritidis are DNA markers, and the penultimate one is a negative control; the first and second ones in the gel graphs of other serotypes of Salmonella and non-Salmonella are DNA markers and positive controls, respectively) .
[0098] The strains and test results are shown in Table 2 below; in the table, "+" in the test result column means positive, and "-" means negative.
[...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com