Single molecule DNA fluorescence signal detection system and detection method for array micro-pores
A fluorescent signal detection and fluorescent signal technology, applied in biochemical equipment and methods, enzymology/microbiology devices, microbial measurement/inspection, etc., can solve the problem of high difficulty in chip processing and preparation, which limits wide application and development, and sequencing High cost and other problems, to achieve the effect of improving detection efficiency and accuracy, increasing utilization rate, and improving fluorescence excitation efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0030] Below, the present invention will be further described in conjunction with the accompanying drawings and specific implementation methods. It should be noted that, under the premise of not conflicting, the various embodiments described below or the technical features can be combined arbitrarily to form new embodiments. .
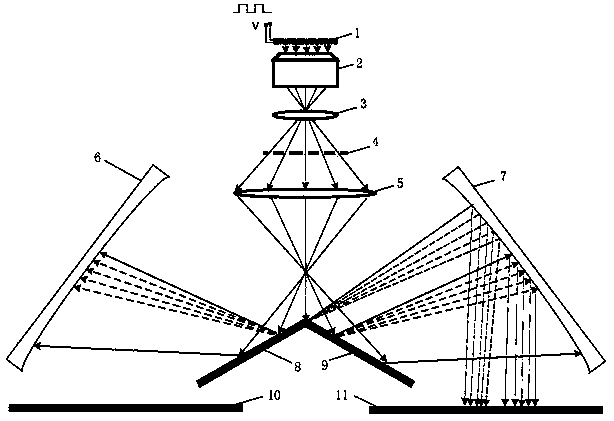
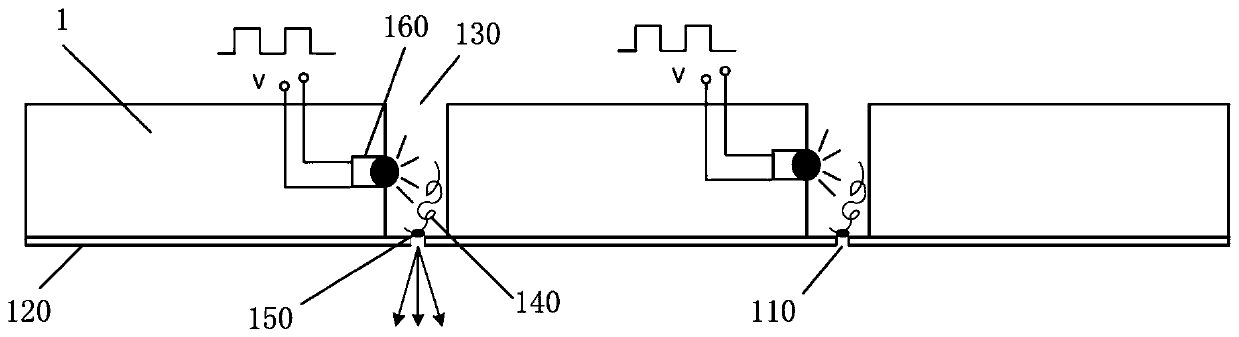
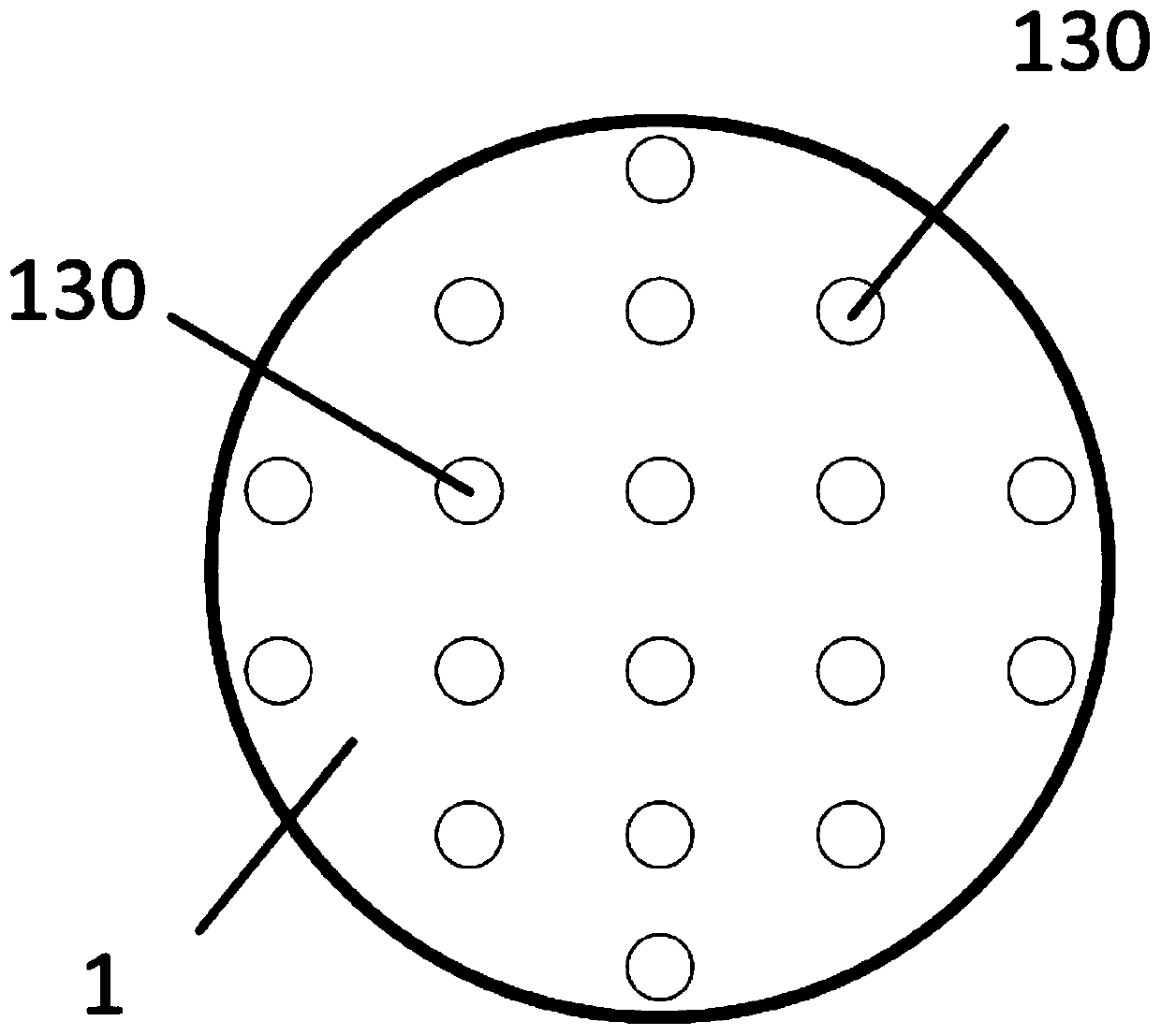
[0031] The invention provides a single-molecule DNA fluorescent signal detection system, such as Figure 1-4 As shown, it includes an array chip 1 and an optical detection structure; wherein, the array chip 1 is arrayed with a number of array microholes 130 and integrated with a number of light-emitting elements, and the light-emitting elements are arranged on the side walls of the array microholes 130 for Illumination, several of the light-emitting elements are electrically connected to a control part, and the array microwells 130 only allow single-stranded DNA molecules 140 to enter;
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com