Method for splitting Cas9 and application
A C-terminal, purpose technology, applied in applications, other methods of inserting foreign genetic materials, botanical equipment and methods, etc., can solve problems such as no longer applicable, reduced splicing efficiency, etc., to achieve the effect of improving editing efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0083] Example 1 Construction method of expression vector
[0084] 1) Resolution method of Cas9 protein
[0085] The Cas9 protein is split into two different amino acid sequences Cas9N protein and Cas9C protein at one of the following positions; the Cas9 protein is spCas9 (D10A) protein, and its amino acid sequence is shown in SEQ ID NO. 1, which encodes the nucleus The nucleotide sequence is shown in SEQ ID NO. 2; the split positions are: between positions 178-179, between positions 203-204, between positions 253-254, between positions 309-310, Between 385-386th, 465-466th, 468-469th, 530-531th, 573-574th, 637-638th, 656th -657, 674-675, 684-685, 713-714, 718-719, 729-730, 769-770 Between the 940-941th position, or the 1005-1006th position. The protein groups obtained according to the resolution method are shown in Table 1 below.
[0086] Table 1 Protein groups obtained from different Cas9 split positions
[0087]
[0088]
[0089] 2) Construction of the nucleic acid construct gr...
Embodiment 2
[0099] Example 2 Functional verification of gene editing
[0100] (1) Detect the function of split-Cas9 containing adenosine deaminase-split-ABE system
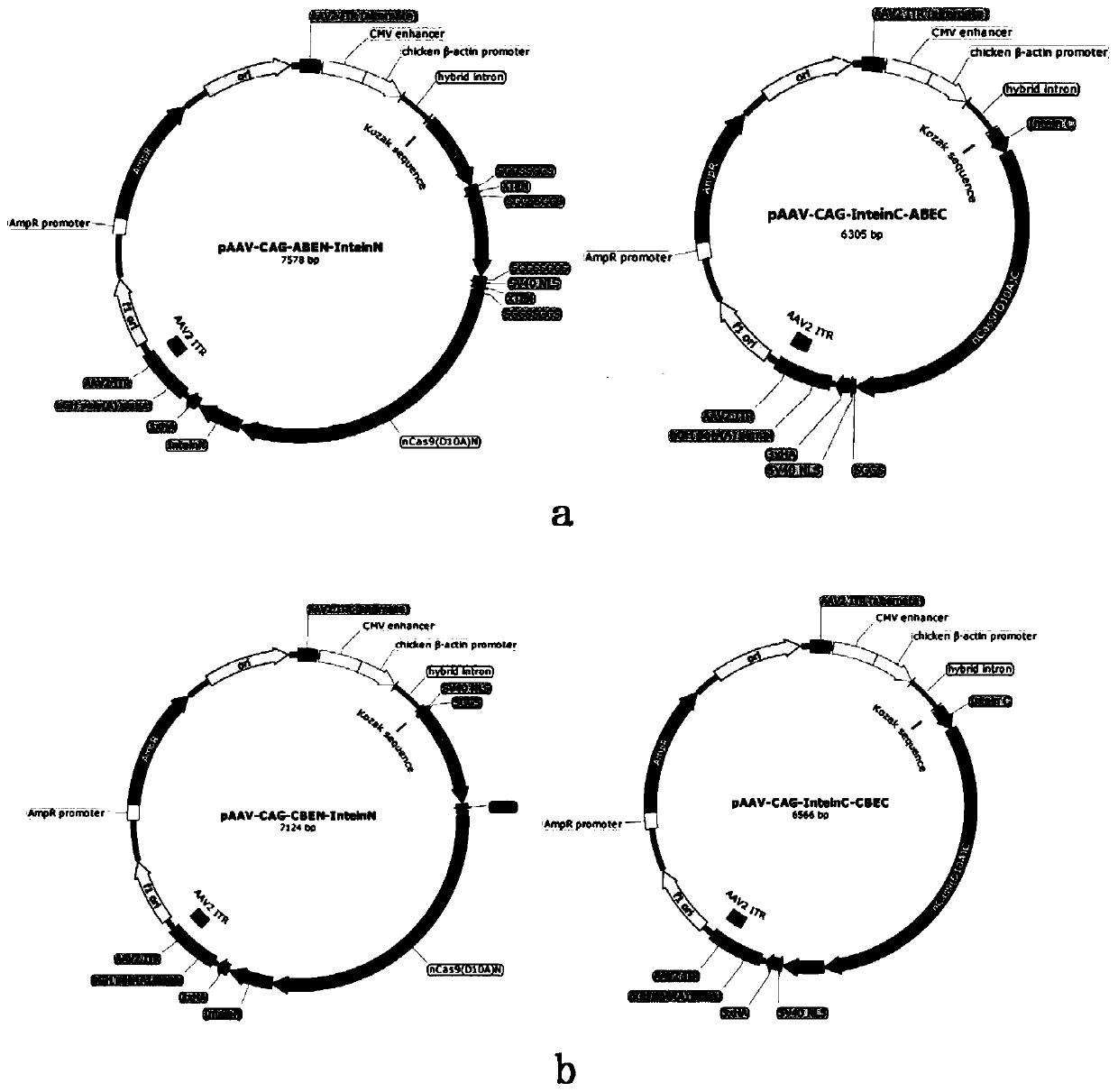
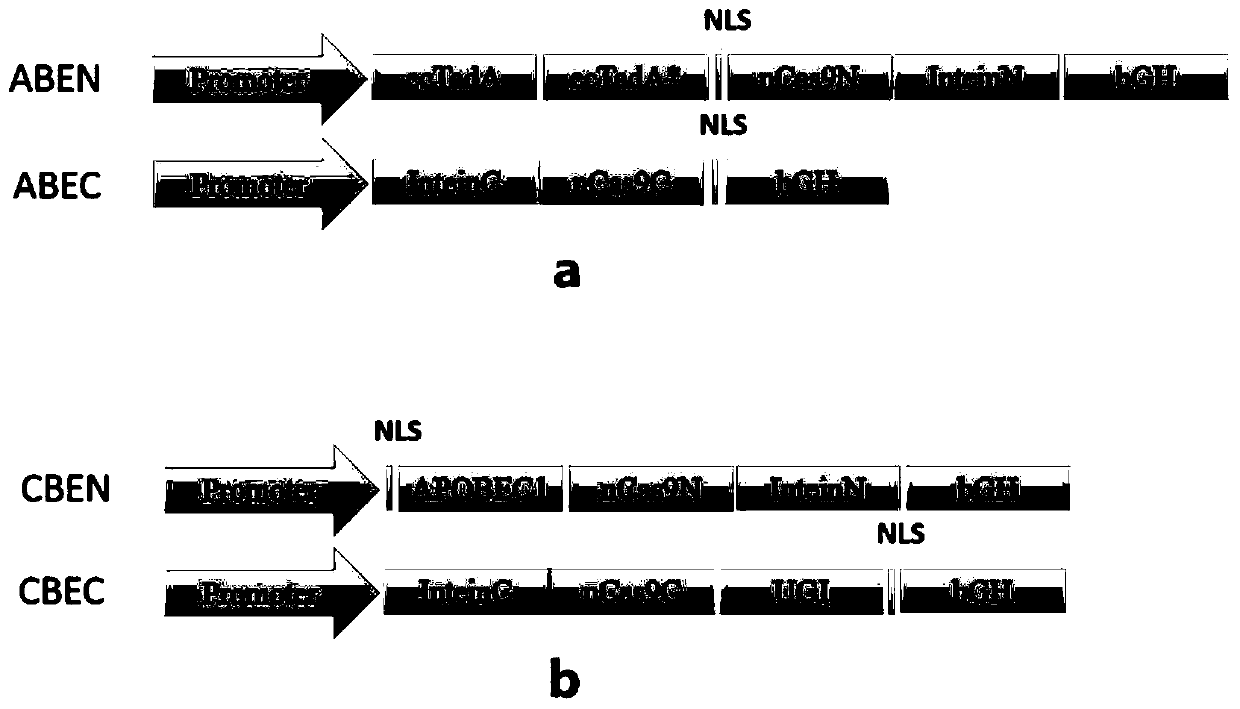
[0101] According to the method of Example 1, the expression vector group (split-ABE system) containing adenosine deaminase was constructed: pAAV-CAG-ABEN-InteinN and pAAV-CAG-InteinC-ABEC. The expression vector group is that the spCas9 protein is split from one of the following positions: between positions 178-179, between positions 203-204, between positions 253-254, and between positions 309-310 , Between 465-466, between 468-469, between 530-531, between 573-574, between 637-638, between 656-657, between Between 674-675, 684-685, 713-714, 718-719, 729-730; the intein is Npu intein, Mxe intein or Rma intein.
[0102] Constructing HEK293T cells containing m1EmGFP stably expressing, the m1EmGFP is the 70th amino acid codon CAG of the EmGFP sequence mutated to TAG, and the EmGFP nucleotide sequence is shown in SEQ ID NO.16. Becau...
Embodiment 3
[0118] Example 3 Activity detection of gene editing by split-ABE system
[0119] The ability of the split-ABE system to perform genome gene editing was further tested; the expression vector set was the spCas9 protein split from one of the following positions: between positions 203-204, between positions 309-310, and positions Between position 573-574, position 674-675, position 684-685; the intein is Npu intein or Rma intein.
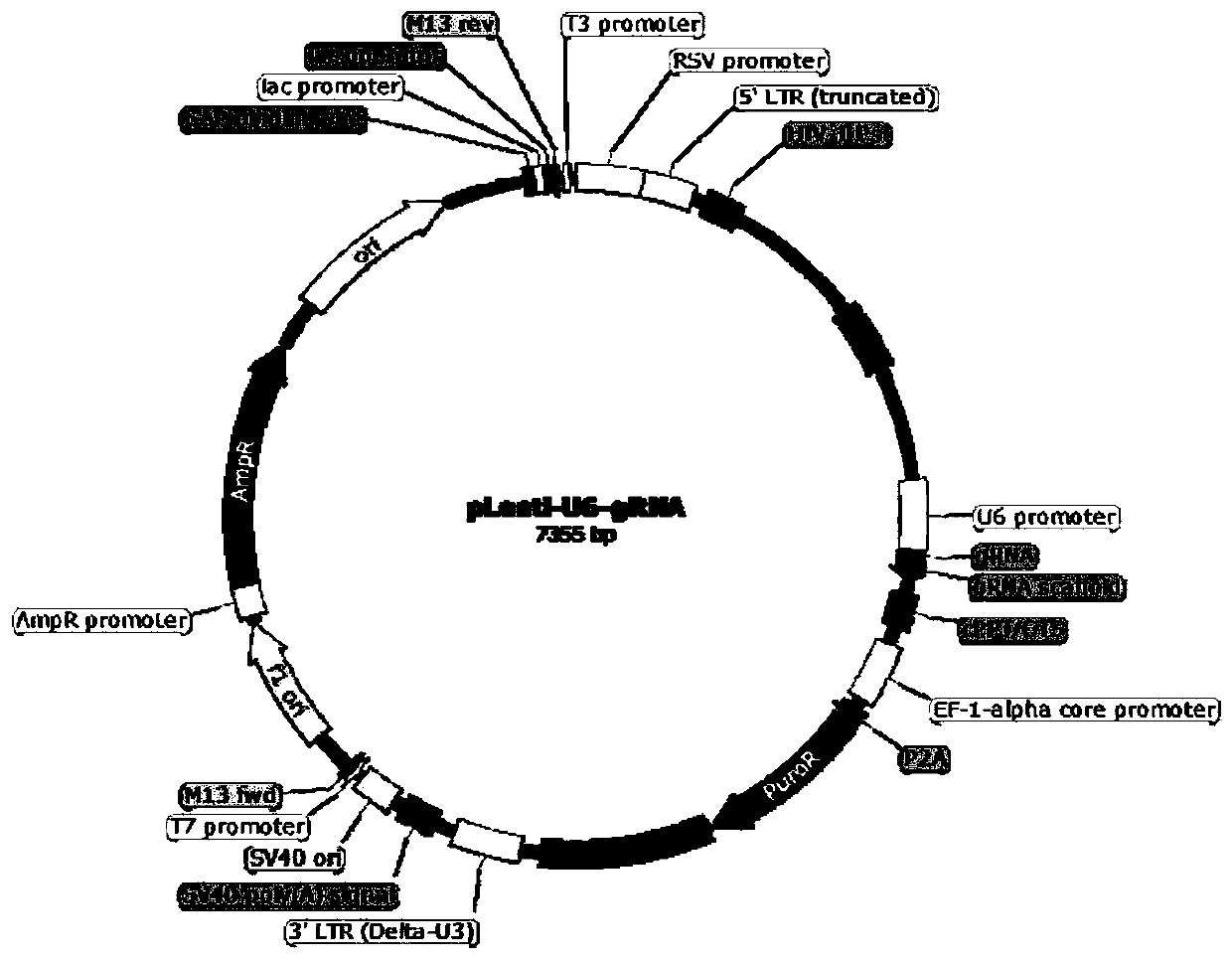
[0120] The gRNA expression vectors pLenti-U6-gRNA-AAVS1, pLenti-U6-gRNA-TERT, and pLenti-U6-gRNA-CCR5 targeting AAVS1, TERT, and CCR5 were constructed according to the method in Example 1. The gRNA sequence of each vector is AAVS1: tccctagtggccccactgtg; TERT: ggtgacaagtgtgatcactt; CCR5: cagccaccctcttttctctg.
[0121] Cultivate HEK293T, HeLa, U2OS to make the cell density about 70%-80%, replace with a new DMEM medium, and divide each cell into an experimental group, a positive control group, and a negative control group (3 multiple wells in each group) , And ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com