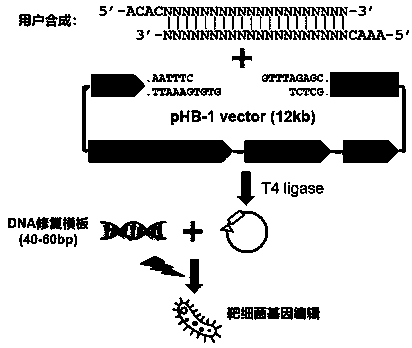
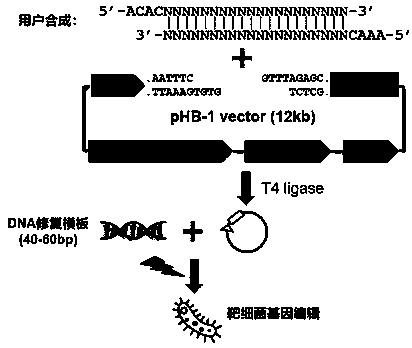
Linearized DNA vector pHB-1 plasmid and test kit prepared from same and used for editing bacterial genomes
A linearization and kit technology, applied in the fields of high-end biology and genetic engineering, can solve the problems of low cost performance, high price, and no kit products, etc., and achieve the effect of precise cut point and low price
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0065] A linearized DNA vector pHB-1 plasmid contains pSC101 replicon, Cas9 gene whose expression is controlled by natural promoter, Gam gene, Beta gene, Exo gene and gRNA whose expression is controlled by IPTG induction; wherein Gam gene, Beta gene and All Exo genes are controlled by the arabinose-inducible promoter ParaB;
[0066] The gene sequence of the linearized DNA vector pHB-1 plasmid is shown in SEQ ID NO:1.
Embodiment 2
[0068] A kit for fast and efficient editing of bacterial genomes, including:
[0069] The linearized DNA vector pHB-1 plasmid (1 ug / uL) described in Example 1;
[0070] High efficiency T4 DNA ligase Mix (2X);
[0071] PCR sequencing primer tL3-seq;
[0072] Experimental control component 1 (point mutation editing of E. coli MlaC gene): primers MlaC-gRNA-F and MlaC-gRNA-R (100uM);
[0073] Experimental control component 2 (point mutation editing of E. coli MlaC gene): DNA repair templates L11R-F and L11R-R (1ug / uL);
[0074] Experimental control component 3: sequencing primer Seq-F1;
[0075] Experimental control component 4: sequencing primer Seq-R1;
[0076] Experimental control component 5: sequencing primer Seq-F2;
[0077] Experimental control component 6: sequencing primer Seq-R2.
[0078] The experimental control component 3 and the experimental control component 4 are used to amplify the DNA fragment centered on the editing site, and the experimental control compo...
Embodiment 3
[0103] A kit for fast and efficient editing of bacterial genomes, including:
[0104] The linearized DNA vector pHB-1 plasmid (1 ug / uL) described in Example 1;
[0105] High efficiency T4 DNA ligase Mix (2X);
[0106] PCR sequencing primer tL3-seq;
[0107] Experimental component 1 (point mutation editing of TolA gene): primers TolA-gRNA-F and TolA-gRNA-R (100uM);
[0108] Experimental component 2 (point mutation editing of TolA gene): DNA repair templates TolA-KO- and TolA-KO-R (1ug / uL);
[0109] Experimental control component 1 (point mutation editing of E. coli MlaC gene): primers MlaC-gRNA-F and MlaC-gRNA-R (100uM);
[0110] Experimental control component 2 (point mutation editing of E. coli MlaC gene): DNA repair templates L11R-F and L11R-R (1ug / uL);
[0111] Experimental component 3: sequencing primer Seq-F1;
[0112] Experimental component 4: sequencing primer Seq-R1;
[0113] Experimental component 5: sequencing primer Seq-F2;
[0114] Experimental component 6: ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com