DNA aptamer binding specifically to tb7.7 and use thereof
A DNA aptamer and specific technology, applied in the field of DNA aptamers, to achieve the effect of eliminating false positive reactions, excellent binding ability and high exclusivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
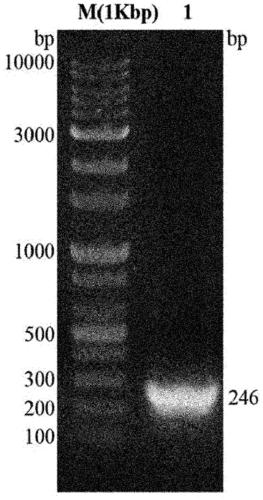
[0049] Embodiment 1.TB7.7 gene cloning
[0050] To amplify the gene for the 7.7kDa tuberculosis-specific antigen (TB7.7), a forward primer including the BamH1 restriction enzyme recognition sequence [5'-CCC GGA TCC ATG AGC GGC CAC GCG TTG3' was used (SEQ ID NO:1)], the reverse primer includes the Xho1 restriction enzyme recognition sequence [5'CCC CCC TCG AGT CAC GGC GGA TCA CCCC3'(SEQ ID NO:2)].
[0051] Genomic DNA of M. tuberculosis H37Rv was used as a template for gene amplification and was amplified by polymerase chain reaction (PCR) using i-pfu polymerase. Each process of PCR is as follows: 1) Incubate at 98°C for 20 seconds, as a process to denature double-stranded DNA into a template; 2) Incubate at 55°C for 20 seconds, as a process for annealing the template with primers; 3) Incubate at 72°C 30 seconds, as a process of extending new strands, and a cycle of repeating these processes 30 times.
[0052] The amplified TB7.7 gene was cloned into a (His)6-tag-containing...
Embodiment 2
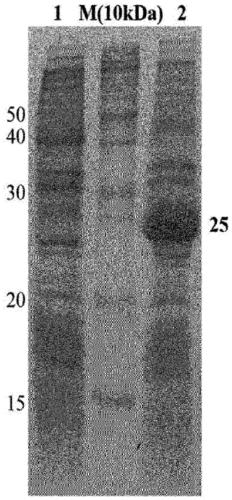
[0053] Expression of embodiment 2.TB7.7 protein
[0054] BL21(DE3) cells transformed with TB7.7 gene were cultured in Luria Bertani(LB) medium and incubated at 37°C until the optical density (OD) at UV 600nm reached 0.563. Thereafter, isopropyl-thio-b-D-galactopyranoside (IPTG) was added to a final concentration of 20 mM to induce the expression of the protein, followed by incubation at 37°C for 4 hours. The expression of the protein was confirmed by SDS PAGE, and the cells were separated from the medium using a centrifuge and washed once with PBS (10 mM sodium phosphate, 150 mM NaCl, pH 8.0) buffer. results, such as Figure 1b As shown, it was confirmed that the recombinant TB7.7 protein was overexpressed.
Embodiment 3
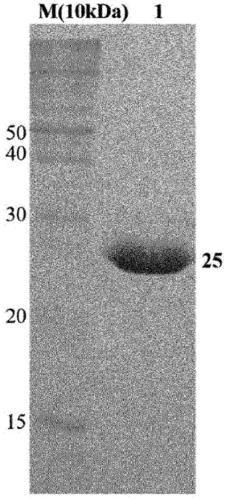
[0055] Purification of embodiment 3.TB7.7 protein
[0056] To purify TB7.7 protein expressed in bacterial cells BL21(DE3) with high purity, cells were lysed in cell lysis buffer (20 mM Tris, 500 mM NaCl, 0.5 mM β-mercaptoethanol, 5% glycerol, pH 8.0), It was then ruptured by ultrasonication for 15 minutes. A centrifuge was used at 18,000 rpm for 40 minutes to separate the protein in an aqueous solution from the cells.
[0057] Furthermore, in order to obtain high-purity proteins, the binding properties between nickel-nitrilotriacetic acid (Ni-NTA) and (His)6-tagged amino acids were utilized. Specifically, as shown in FIG. 1 , a Ni-NTA column was connected to a fast protein liquid chromatography (FPLC) system, and TB7.7 in an aqueous solution state was flowed into the column, thereby allowing TB7.7 to bind thereto. Since the (His)6-tag of the protein bound to the Ni-NTA column competes with the imidazole compound to separate the target protein from the column, the elution b...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com