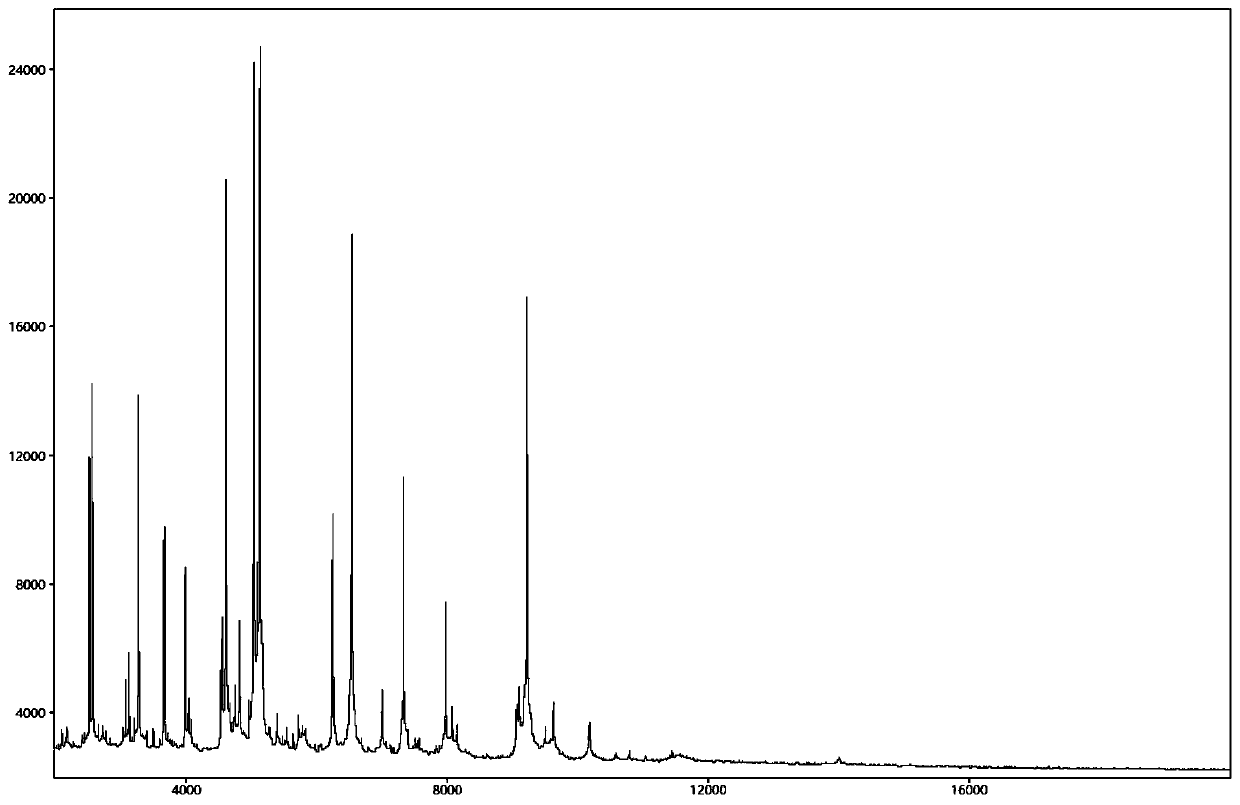
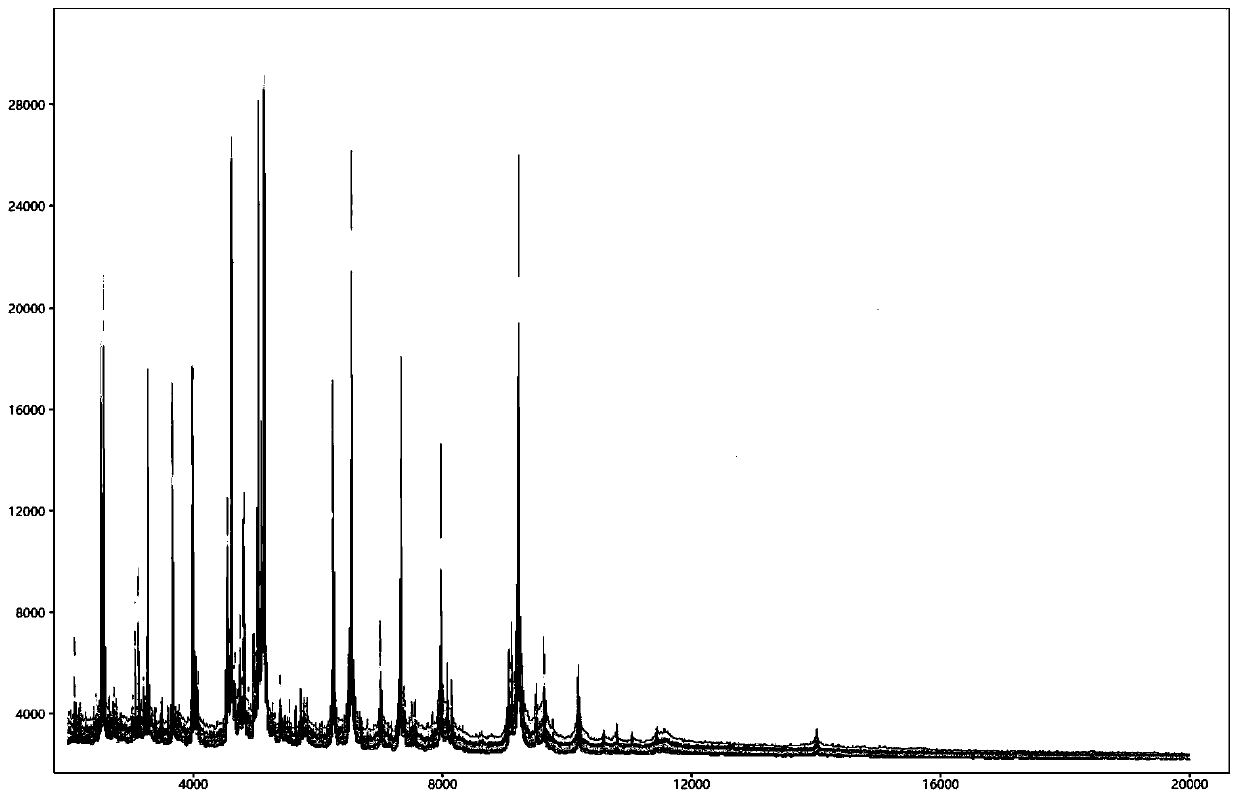
Database establishment method and identification method of BALDI-TOF MS strain
A technique for establishing a method and Bartonella, which is applied in the field of microbial mass spectrometry detection, can solve problems such as impossibility and limitation of technology application and development, and achieve the effects of saving operation steps, shortening identification time, and easy operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1,200、21MALDI-TOF MS ,11ATCC(1 no. 3)。 Embodiment 2
[0078] In Example 1, 200 strains and 21 Bartonella strains were selected for establishing a MALDI-TOF MS identification database, which contained 11 Bartonella ATCC standard bacteria (column 3 in Table 1). In Example 2, 173 strains of Bartonella wild isolates were used to evaluate the identification reliability of the established database (column 4 in Table 1). The experimental strains used were provided by the Vector Biology Control Office of the Institute of Infectious Diseases Control, Chinese Center for Disease Control and Prevention.
[0079] Table 1: Strain information used to establish and evaluate the Bartonella MALDI-TOF-MS database
[0080]
[0081] Note: 1. * is pathogenic Bartonella; 2. "This laboratory" is the vector biological control room of the Chinese Center for Disease Control and Prevention; the Bartonella strains of "this laboratory" can be obtained from public strains It can be obtained from the preservation center or isolated using existing technology...
Embodiment 1
[0083] The present embodiment is the establishment method of the Bartonella database utilizing MALDI-TOF MS, and the method comprises the following steps:
[0084] (1) Cultivation and identification of Bartonella strains
[0085] Among the Bartonella strains (alternative strains) in Table 1, the culture conditions for Bacilliformis (B.bacilliformis) are: at 28°C, 5% CO 2 In the incubator, resuscitate culture on TSA medium of 10% defibrinated sheep blood. The culture conditions for other Bartonella strains are: at 37°C, 5% CO 2 In the incubator, cultivate under the environment of 70% humidity, and revive the culture on the TSA medium of 5% defibrated sheep blood. Harvest the fresh bacteria from the 4th to 6th day of culture according to the growth rate of the colony.
[0086] Using primers BhCS.781p-BhCS.1137n to amplify the specific gene of Bartonella genus citrate synthase gene (gltA) for taxonomic identification [6] . The PCR primers used for identification are as follo...
Embodiment 2
[0107] Embodiment 2: wild strain actual measurement
[0108] This embodiment utilizes the Bartonella standard database of MALDI-TOF MS established in Example 1 to identify whether the strains to be tested (14 species, 173 strains in total) are Bartonella. The method comprises the following steps:
[0109] (1) Cultivation and identification of Bartonella strains:
[0110] Resuscitate and culture the strain to be tested to obtain fresh bacteria; then perform PCR reaction and sequence on the fresh bacteria to determine whether it is Bartonella. The specific operation method and parameters are as described in Example 1.
[0111] (2) Sample pretreatment:
[0112] The identified fresh bacteria were pretreated to obtain a sample protein solution. The specific operation method and parameters are as described in Example 1.
[0113] (3) Target plate preparation:
[0114] Add the sample protein solution dropwise to the target position to prepare a target plate. The specific operati...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com