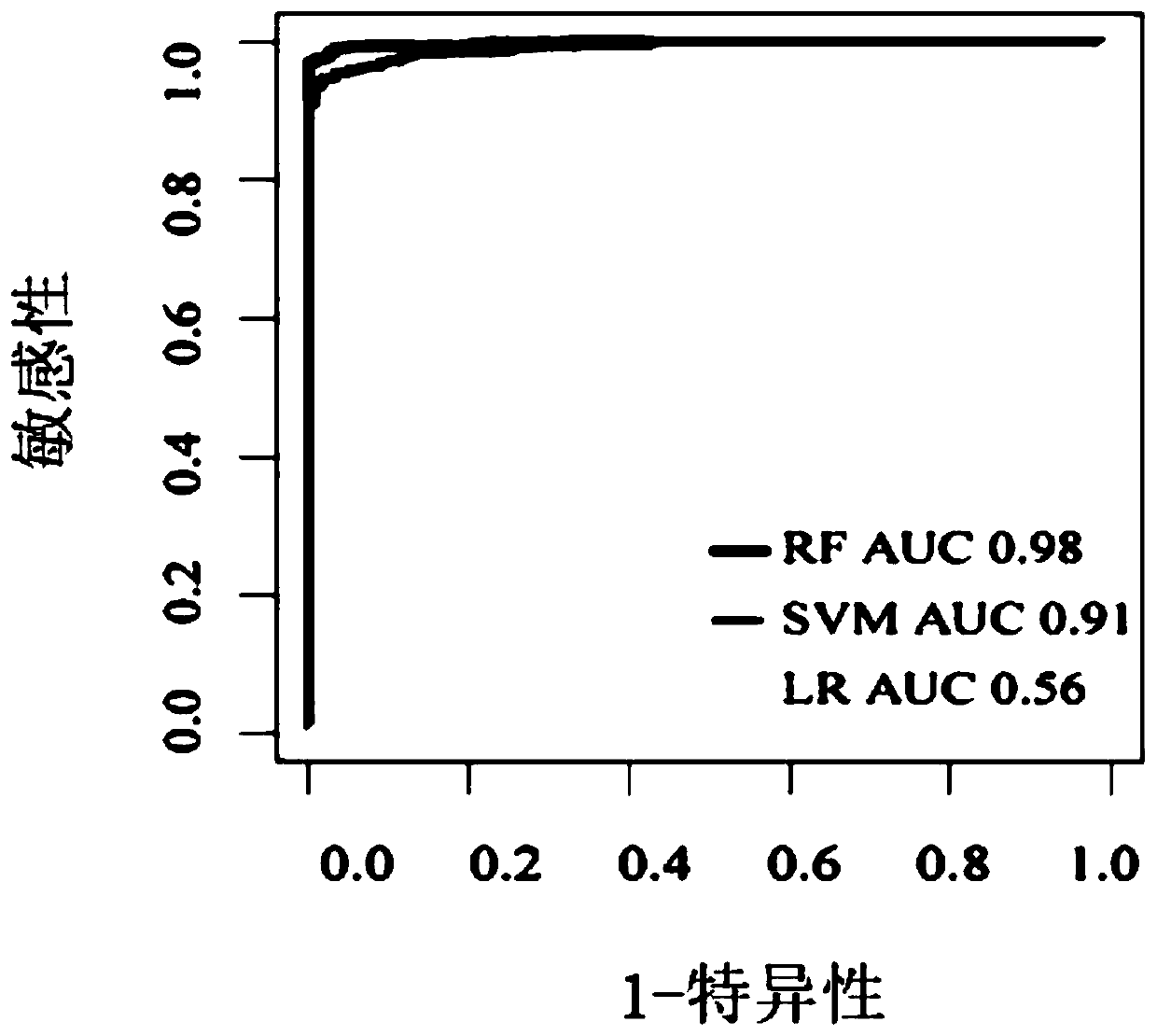
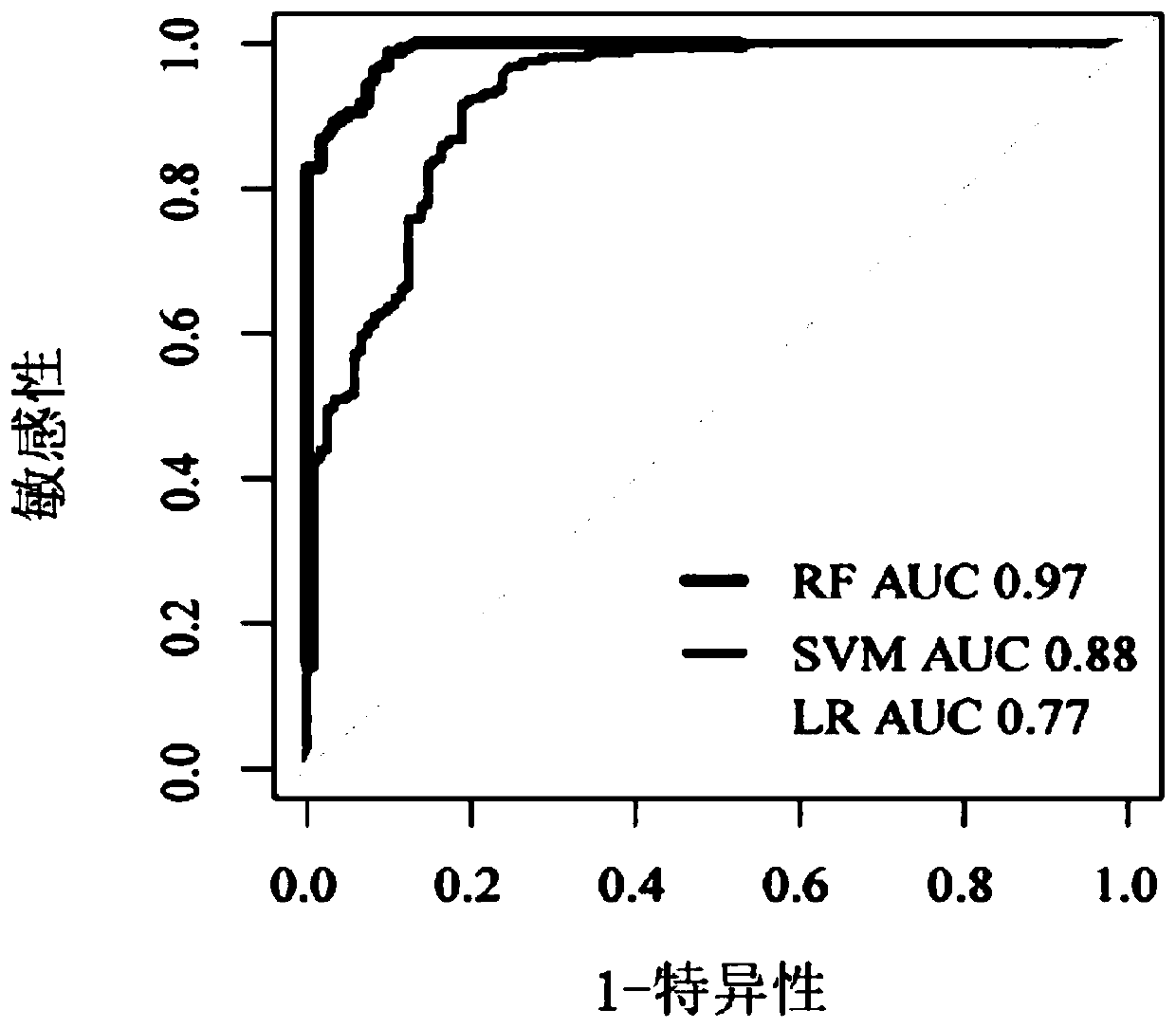
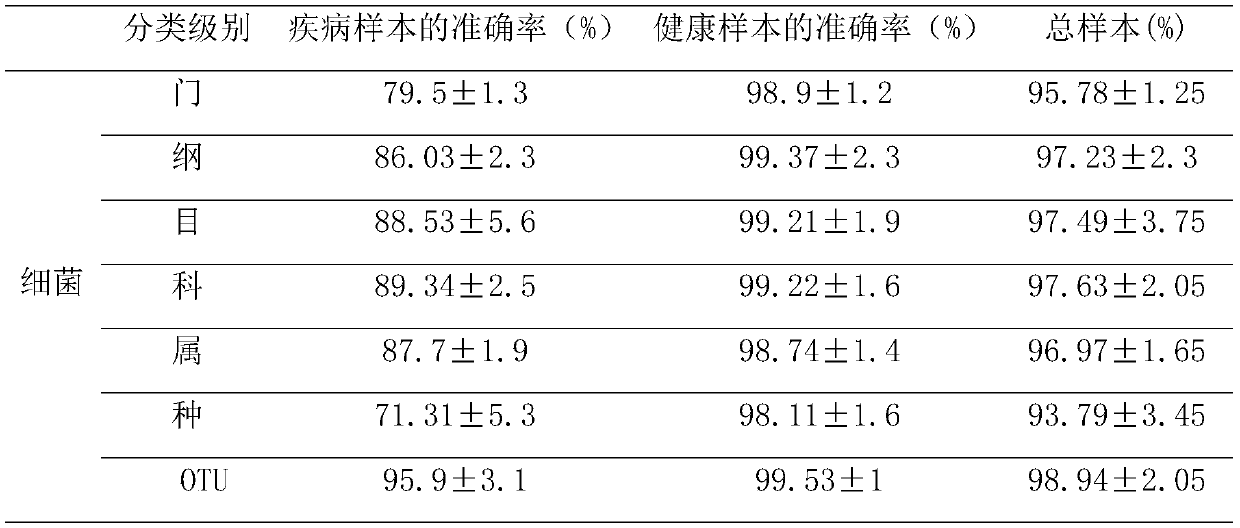
Wilt disease prediction model based on high-throughput sequencing data and application
A technology for sequencing data and prediction models, applied in sequence analysis, instruments, biological systems, etc., can solve the problems of rising incidence and inability to predict diseases by pathogenic bacteria, and achieve the effect of simple application operation and reliable prediction results.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0033] A Fusarium wilt incidence prediction model based on high-throughput sequencing data, comprising the following steps:
[0034] 1. Data collection
[0035] Based on the published research, use the keywords Fusarium wilt community (Fusarium wilt community) or Fusarium wilt structure (Fusarium wilt structure) or Fusarium wilt microbiome (Fusarium wilt microbiome) to search for research papers related to fungal wilt and microbial communities from Google Scholar, and get Sequencing sample accession numbers for Fusarium wilt-associated soil microbial communities. Most of the original data of related studies are stored in NCBI, and some are stored in DDBJ and European Nucleic Acid Database, from which sequencing data information can be obtained.
[0036] At present, NCBI stores a large amount of data that has not yet been published. The grouping of these data and their sampling information are often described very clearly. You can search for the keywords Fusarium wiltcommunity...
Embodiment 2
[0061] To verify the accuracy of the bacterial model, 13 independent bacterial datasets from the NCBI SRA database were analyzed next, including 26 fusarium wilt-onset soil samples and 321 healthy soil samples. For all collected samples, the average accuracy rate of the bacterial model was 94.46%, the accuracy rate of the diseased sample was 91.75%, and the accuracy rate of the healthy sample was 96.45% (see Table 3).
[0062] At the same time, 6 independent fungal data sets from the NCBI SRA database, including 21 soil samples with Fusarium wilt disease and 144 healthy soil samples, were used to validate the fungal model. The average accuracy of the fungal model for all collected samples was 93.05%, the accuracy for diseased samples was 91.67%, and the accuracy for healthy samples was 95.25% (see Table 4).
[0063] Table 3 Bacterial model prediction accuracy of NCBI dataset
[0064]
[0065] Table 4 Prediction accuracy of fungal model in NCBI dataset
[0066]
[0067]...
Embodiment 3
[0069] Soil sample collection: In late April 2019, field soil samples of 4 different crops of banana, cucumber, watermelon and lily in Hainan, Guangdong, Beijing and Jiangsu were collected. For the soil collection of diseases and insect pests, plots that had been continuously cropped and suffered from Fusarium wilt for at least 5 years were selected, while newly planted plots without Fusarium wilt were selected for healthy soil collection. In order to avoid the differences caused by geographical factors, the healthy ones are selected near the diseased areas. For each soil sample, 20 random soil cores (diameter 5cm×20cm) were taken from the 0-20cm soil layer, and each plot was sampled in S-shape to form a soil sample. Freshly collected soil was screened through a 2-mm sieve to remove plant debris and possible small animals, and then stored at -80°C prior to DNA extraction.
[0070] DNA extraction, gene amplification and sequencing: (1) Power Lyzer PowerSoil DNA Isolation Kit (...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com