A method for nucleic acid aptamer to identify target protein CD63 and its application in overcoming vemurafenib resistance in melanoma
A nucleic acid aptamer and target protein technology, which can be used in anti-tumor drugs, medical preparations containing active ingredients, drug combinations, etc., can solve the problem of unknown application of anti-vemurafenib, and achieve the effect of enhancing sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
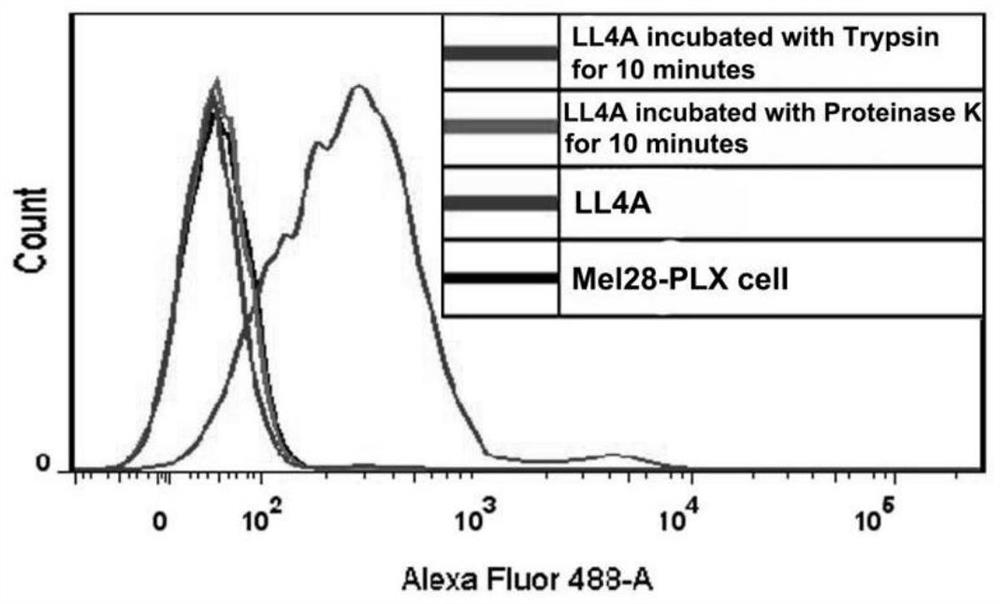
[0055] Example 1: Target type identification of nucleic acid aptamer LL4A
[0056] The experimental steps are as follows:
[0057] (1) Preparation of ssDNA: Take 50μM nucleic acid aptamer LL4A and library (Library), denature at 95°C for 5min, refold on ice for 10min, centrifuge at 5000rpm at 4°C for 3min, add Binding buffer to make the DNA concentration 250nM, place on ice stand-by.
[0058] ssDNA sequence (LL4A):
[0059] GCTGGACTCACCTCGACCAGAGCCATTGGGTTTCCTAGGAAATAGGGCCTTTACTATGAGCGAGCCTGGCG;
[0060] (2) Cell preparation: culture Mel28-PLX cells in four dishes to the logarithmic growth phase, wash twice with 2mL DPBS, add 200μl 0.25% trypsin and 200μl 0.1mg / ml proteinase K to two of the dishes to digest at room temperature for 10min , the other two dishes were digested with 0.2% EDTA under the same conditions, and then 500 μl of complete medium was added to terminate the digestion, centrifuged at 800 rpm for 4 min, and the supernatant was discarded. Wash twice with Wash...
Embodiment 2
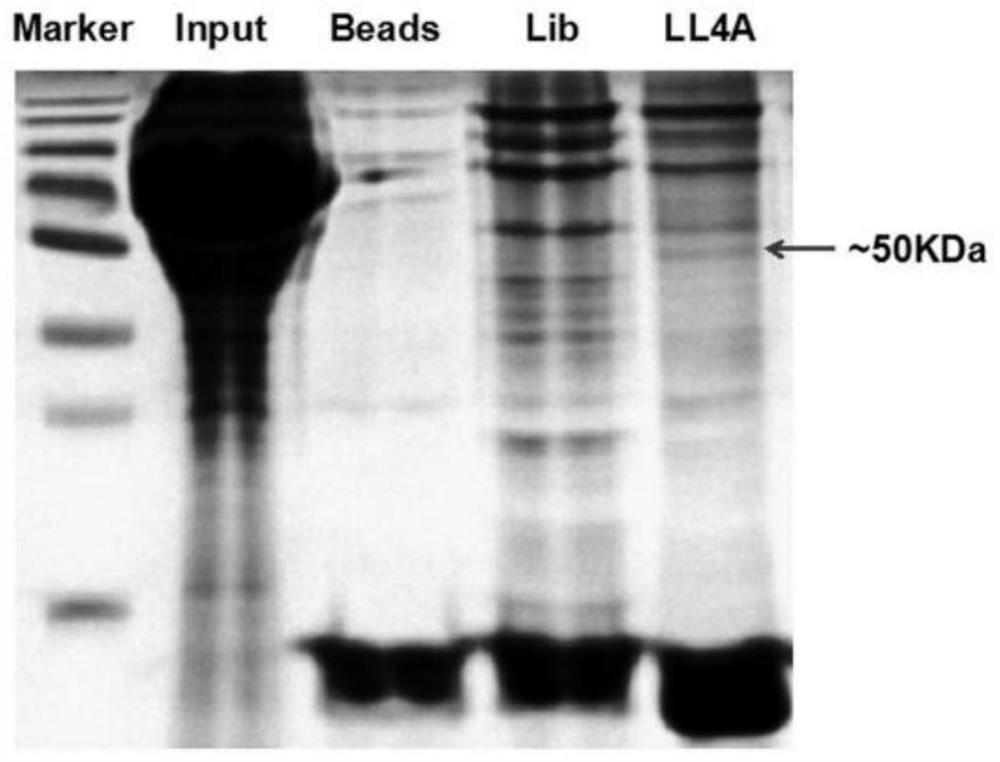
[0062] Example 2: Nucleic acid aptamer target mass spectrometric identification
[0063] 1) Extraction of cell membrane proteins
[0064] (1) Inoculate Mel28-PLX cells in 10 10cm culture dishes and culture them to the logarithmic phase;
[0065] (2) Discard the old culture medium and wash it twice with DPBS;
[0066] (3) After digestion with enzyme-free digestion solution, collect the cells in a 15mL centrifuge tube with DPBS;
[0067] (4) Centrifuge and wash with Washingbuffer, discard the supernatant;
[0068] (5) Add a corresponding amount of hypotonic buffer (300 μL / dish) into a 15 mL centrifuge tube, vortex and mix well, and shake at 4° C. for 30 min. 4000rpm, centrifuge for 10min and discard the supernatant. Then wash with hypotonic buffer 3 times;
[0069] (6) Add a corresponding amount of membrane protein lysate (200 μL / dish) to the centrifuge tube, shake and mix well, lyse at 4°C for 30 minutes, centrifuge to retain the supernatant, 4000 rpm, 4°C, 10 minutes. Th...
Embodiment 3
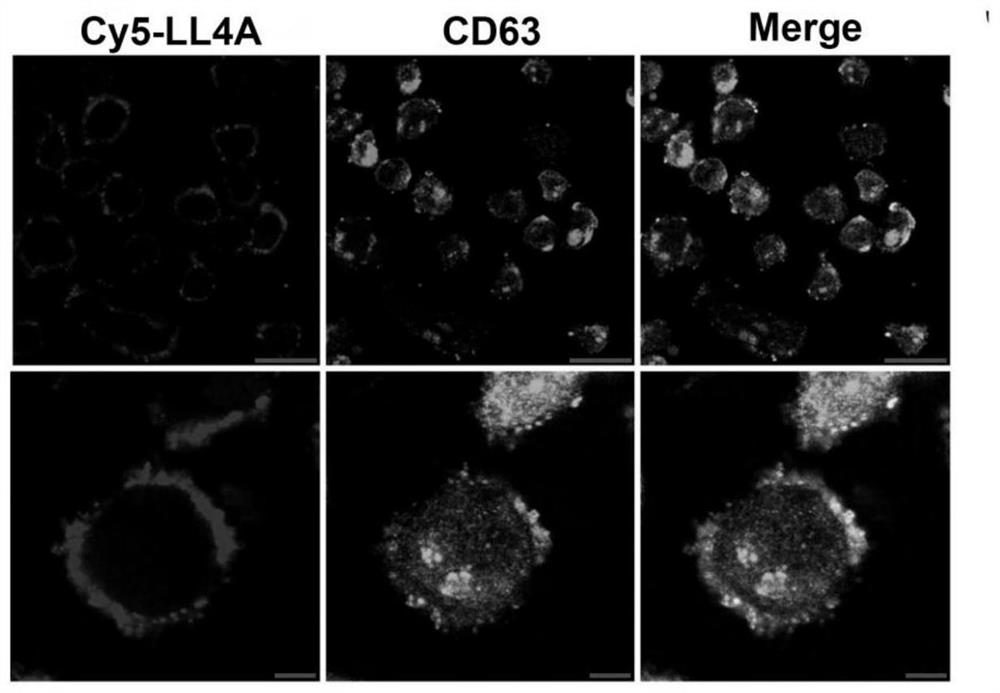
[0086] Example 3: Co-localization of LL4A and CD63 proteins
[0087] Laser confocal fluorescence microscopy can visually display the fluorescent signals on the cell surface. In this experiment, the colocalization experiment of nucleic acid aptamer and CD63 protein was carried out by laser confocal fluorescence microscopy, which further proved that the target of LL4A is CD63 protein.
[0088] The specific experimental steps are as follows:
[0089](1) Cell preparation: Mel28-PLX cells were connected to the optical culture dish 24 hours in advance, so that the confluence of the cells reached 90%-95% when used, washed three times with Washing buffer, blocked with 1% BSA for 30 minutes, and aspirated after sealing. abandoned.
[0090] (2) Preparation of LL4A and antibody: 10 μL of FITC-labeled CD63 antibody and 50 pmol of LL4A labeled with Cy5 fluorophore at the 5′ end were added to a 1.5 mL EP tube containing 200 μL of Bindingbuffer.
[0091] (3) Incubation of cells with LL4A a...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com