Intestinal flora metagenome database construction method and device and intestinal flora metagenome database analysis method and device
A technology of metagenomic and intestinal flora, applied in the field of building gut flora metagenomic database, can solve the problems of difficult groups, difficult to elucidate, and no gut flora metagenomic data database, etc., to achieve accurate and reliable information, It is beneficial to management and the interaction of database data
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0078] Glossary:
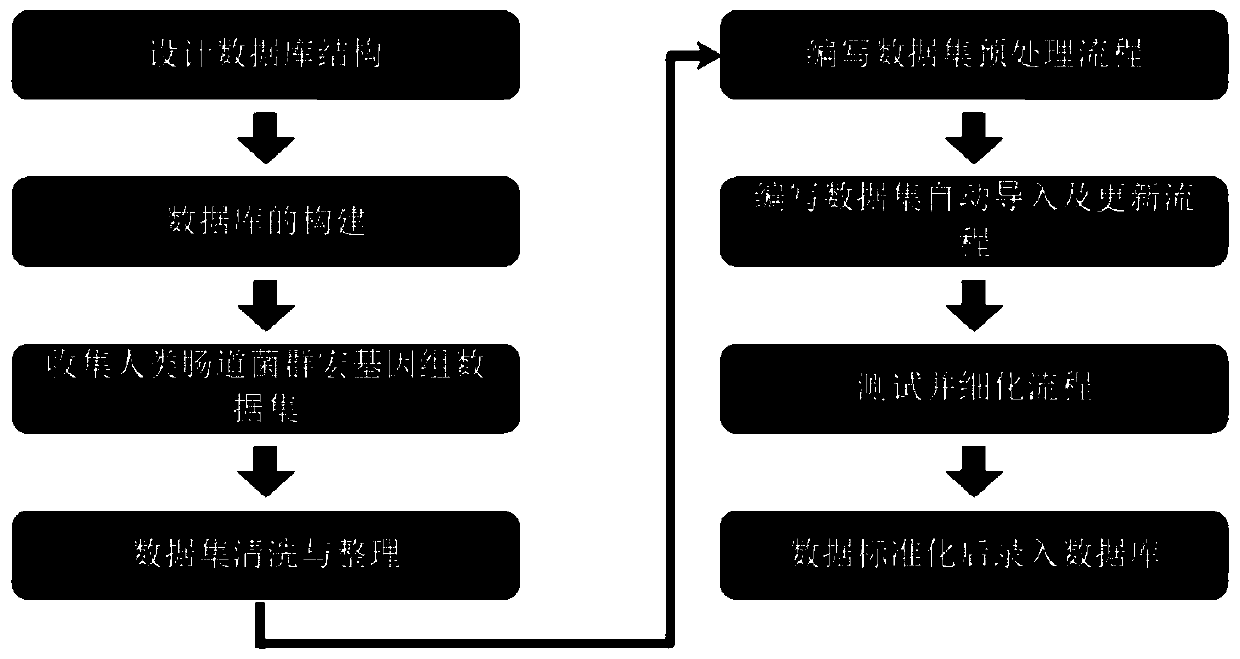
[0079] Sra2HMGA is a package file of database data preprocessing process written by Perl, which contains standard input folder input, output folder output, folder for running process (bin), process description file (readme), etc.
[0080] Phenotype.xls is a standardized sample phenotype file, and the phenotype information that does not exist in the sample is uniformly processed as NA.
[0081] kneaddata_read_count_table.tsv is the data filtering and quality control information obtained after the Fastq file analyzed by the open source software Kneaddata and then processed by the Biobakery process.
[0082] merged_abundance_table_species.txt is the species abundance information file obtained after analyzing the input Fastq file by the open source software Metaphlan2. Specifically, this file belongs to the species abundance information at the "species" level, and the species abundance information is based on Relative abundance after sample normalization.
[0...
Embodiment 2
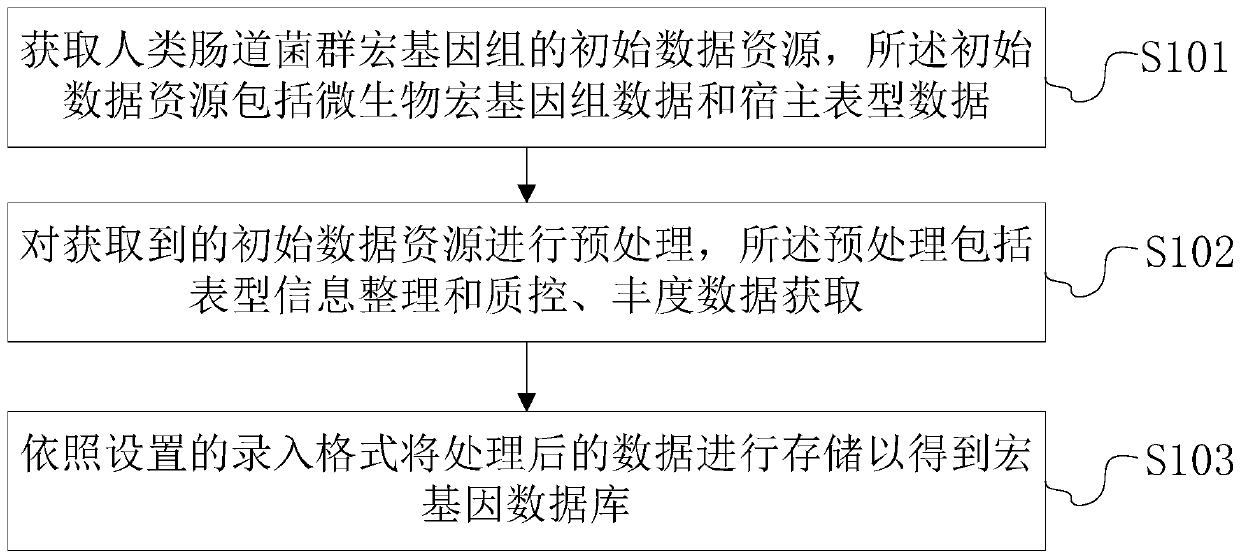
[0117] Such as Figure 5 and Figure 6 As shown, this embodiment provides a system for constructing an intestinal flora metagenomic database, including the following modules:
[0118] Acquisition module: used to obtain the initial data resources of human intestinal flora metagenomics, the initial data resources include microbial metagenomic data and host phenotype data;
[0119] Preprocessing module: used to preprocess the acquired initial data resources, the preprocessing includes phenotypic information sorting and quality control, and abundance data acquisition;
[0120] Storage module: used to store the processed data according to the set input format to obtain the macrogene database.
[0121]The database construction method includes constructing internal storage database and public storage database, constructing database query module, constructing database data interaction system and constructing database data analysis system. The data interaction system includes data a...
Embodiment 3
[0129] Embodiment 3 discloses a computer-readable storage medium, which is used to store a program, and when the program is executed by a processor, the method for constructing an intestinal flora metagenomic database of Embodiment 1 is implemented.
[0130] Of course, a storage medium containing computer-executable instructions provided by an embodiment of the present invention, the computer-executable instructions are not limited to the above-mentioned method operations, and can also perform related operations in the methods provided by any embodiment of the present invention .
[0131] Through the above description about the implementation mode, those skilled in the art can clearly understand that the present invention can be realized by means of software and necessary general-purpose hardware, and of course it can also be realized by hardware, but in many cases the former is a better implementation mode . Based on this understanding, the essence of the technical solution ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com