Kit for quantitatively detecting non-structural protein residues in foot-and-mouth disease inactivated antigen and application thereof
An inactivated antigen quantitative detection technology, applied in the biological field, can solve the problems of time-consuming and costly, and achieve the effect of high sensitivity and accurate calculation results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0054] Example 1 Preparation of 3ABC monoclonal antibody and polyclonal antibody
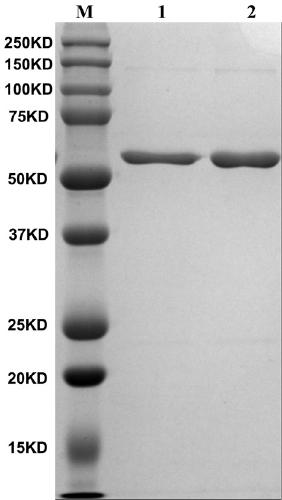
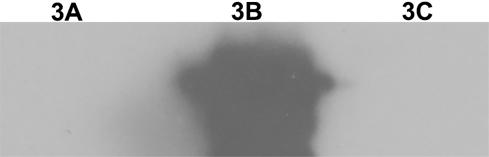
[0055] The foot-and-mouth disease NSP 3ABC of prokaryotic expression is purified ( figure 1 ), and perform gradient dialysis refolding on it, and then prepare monoclonal antibody by 3ABC immunized mice after refolding, subcloning the positive hybridoma cells three times by limiting dilution method, and screening out highly reactive monoclonal cells, Then the screened cells were injected into the peritoneal cavity of mice to prepare ascites. Specific operation process: Inject 0.5ml of Freund's incomplete adjuvant into the peritoneal cavity of female Balb / c mice, and inject about 7.0×105 hybridoma cells into the peritoneal cavity 3 days later. After 9-10 days, ascitic fluid was collected and purified. Verification by Western blot showed that the monoclonal antibody recognized 3B protein, and WB results also showed that the monoclonal antibody recognized a linear epitope ( figure 2 ).
[0056]...
Embodiment 2
[0057] Example 2 Optimization of the CLIA method for detecting foot-and-mouth disease non-structural proteins and establishment of a standard curve
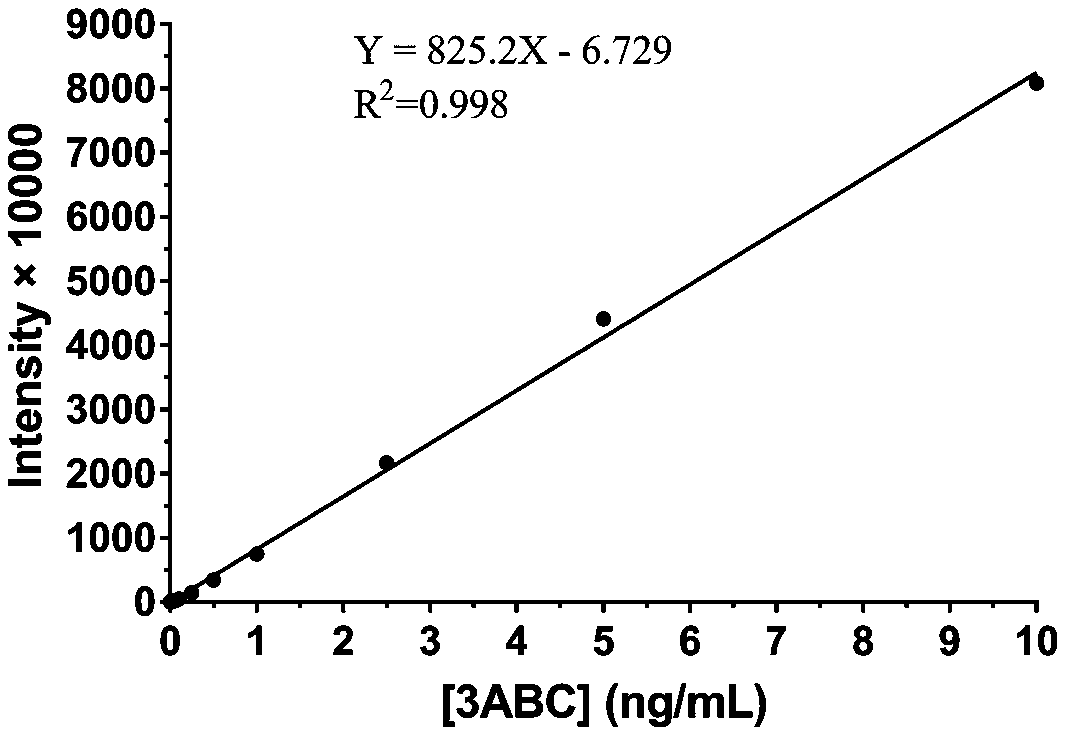
[0058] Dilute the 3ABC polyclonal antibody to 0.5 μg / ml as the capture antibody, dilute the 3B monoclonal antibody-HRP 1:10,000, 1:30,000, and 1:100,000 times as the detection antibody, and dilute the prokaryotic expressed foot-and-mouth disease NSP 3ABC to 500 with PBST, 250, 100, 50, 25, 10, 5, 2.5, 1, 0.5, 0.25, 0.1, 0.05, 0.025, 0.01, 0 ng / ml were used as standard antigens. Considering economic factors and detection range, the optimal coating concentration of capture antibody was finally determined to be 0.5 μg / ml, the optimal dilution of detection antibody was 1:10,000, and the optimal reaction time was 30 min, and a standard curve was established. Such as image 3 As shown, when the concentration of 3ABC is 0.1-10ng / ml, the measured chemiluminescence intensity has a linear relationship with the concentration of 3ABC, and t...
Embodiment 3
[0059] Embodiment 3 Determination of the optimal dilution factor of the foot-and-mouth disease inactivated antigen sample to be inspected
[0060] Since some antigens contain some components that affect the antigen-antibody reaction, the sample to be tested is diluted 1:2, 1:4, 1:8 and compared with the chemiluminescence value (CLIA value) of the original solution to determine the optimal dilution Spend. By comparing 8 antigens purified for different times (not one strain was used), it can be found that after serial dilution of some antigens, the CLIA value decreases proportionally (such as a, b, c, d); while some antigens are serially diluted After that, the CLIA value began to decrease proportionally from 1:4-1:8 (such as 1, 2); and the amount of NSPs contained in the antigen purified twice was already lower than the detection limit of this method, so after serial dilution, CLIA Values vary little (eg I, II). Through this test, the optimal dilution of the antigen was fin...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com