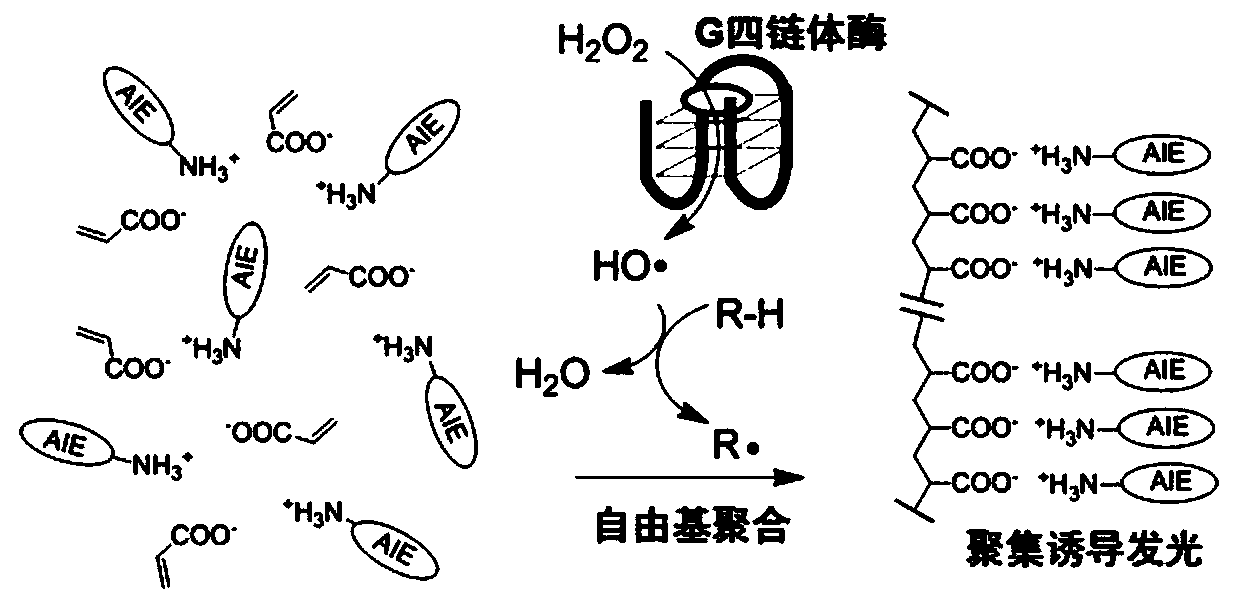
Fluorescence detection signal amplification system
A signal amplification and fluorescence detection technology, applied in the field of fluorescence detection signal method system, can solve problems such as false positives, sequence specificity is not absolute, and samples are easily contaminated.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 3
[0143] Embodiment 3, olefinic fluorescent monomers are applied to the detection of thrombin.
[0144] Configure 25mM HEPES buffer, pH=8, containing: KCl, 1mM; heme, concentration 0.001mM; acetylacetone, 0.001mM; hydrogen peroxide, 0.001mM; DNA of G quadruplex sequence, sequence: 5'- GGTTGGTGTGGTTGG-3'(SEQ ID NO.2), concentration 0.0005mM, the sequence is also a thrombin aptamer sequence; fluorescent monomer, 0.1mM, the fluorescent monomer is:
[0145]
[0146] The above solution can be used for the detection of thrombin. Add 0.0005mM thrombin and mix well. After 20 minutes, the fluorescence is enhanced ten times.
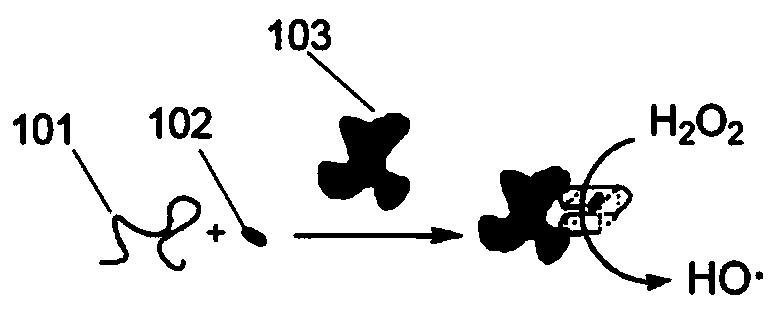
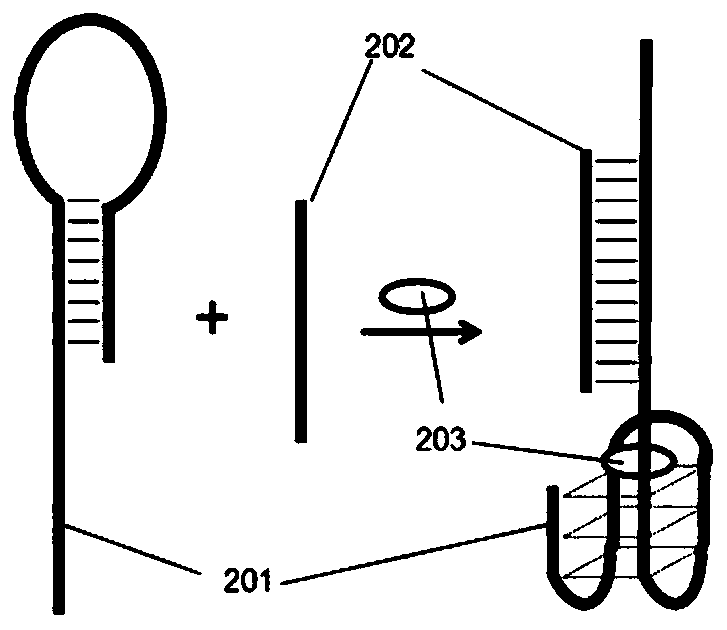
[0147] The detection principle is attached figure 2Shown: In the absence of thrombin 103, DNA sequence 101 is difficult to form a stable G quadruplex, so it cannot bind heme. When there is thrombin, DNA sequence 101 combines with thrombin 103 to form a stable G quadruplex The body structure, combined with heme 102, can catalyze hydrogen peroxide to generate...
Embodiment 4
[0160] Implementation example 4, mercaptoenyne click reaction applied to DNA detection.
[0161] The mercaptoenyne click reaction is slightly different from the olefin polymerization reaction. The mercapto group acts as a free radical transfer during the reaction, so a very small amount of free radicals can trigger the reaction of a large number of functional groups until the mercapto group is consumed.
[0162] Configure 25mM HEPES buffer, pH=7, containing: 100mM KCl; complexes, concentration 0.001mM; mercapto compound monomers, 0.05mM; olefin fluorescent monomers, 0.05mM; DNA probe sequence: 5'-GGGTAGGGCGGGTTGGGAGTTAGCACCCAACCC- 3' (SEQ ID NO. 3), concentration 0.0005 mM.
[0163] Among them, the complex is manganese phthalocyanine, the structure is
[0164] The monomer structure of mercapto compound is:
[0165] Fluorescent monomers are:
[0166] The above solution can be used for target DNA detection. The target sequence: 5'-TGGGTGCTAACT-3' (SEQ ID NO.4), is com...
Embodiment 5
[0183] Example 5, mercaptoenyne click reaction applied to DNA detection.
[0184] Configure 25mM HEPES buffer solution, pH=7, containing: 100mM KCl; heme, concentration 0.001m; hydrogen peroxide, concentration 0.0005mM; fluorescent monomer, 0.05mM;
[0185] DNA probe sequence 1: 5'-ATGACTATCTTTAAT GGGTAGGG-3' (SEQ ID NO.6), concentration 0.001 mM; DNA probe sequence 2: 5'-GGGTTGGG CGTATGGAAAATGAG-3' (SEQ ID NO.7), concentration 0.001 mM.
[0186] Wherein the fluorescent monomer molecule is:
[0187] The above solution can be used for target DNA detection, the detectable target sequence is: 5'-CTCATTTTCCATACATTAAAGATAGTCAT-3' (SEQ ID NO.8), its 5' terminal sequence CTCATTTTCCATACA can be formed with the 3' terminal sequence CGTATGGAAAATGAG of probe sequence 2 Complementary pairing; its 3' terminal sequence TTAAAGATAGTCAT can form a complementary pairing with the 5' terminal sequence ATGACTATCTTTAAT of probe sequence 1.
[0188] Add 0.001 mM target sequence to the solutio...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com