Molecular characterization based on prediction of protein affinity and application thereof
A protein and affinity technology, applied in the analysis of two-dimensional or three-dimensional molecular structure, molecular design, instruments, etc., can solve the problems of incomplete, false negative and incomplete links between molecules and biological activities, and achieve superior skeleton transition performance , Build robust and accurate effects with a wide range of applications
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
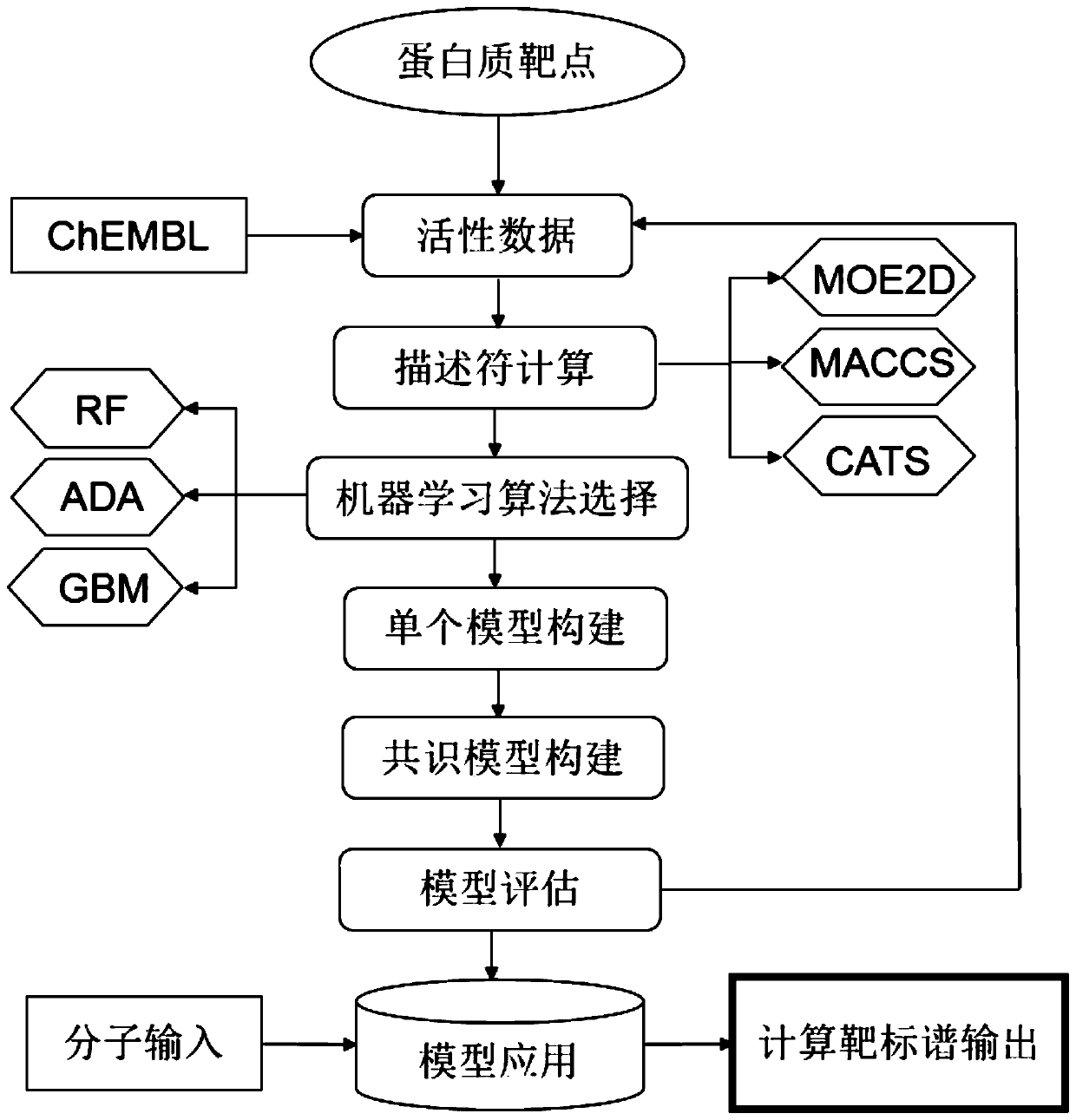
[0064] A computational target spectrum, its construction process see figure 1 , the specific construction steps are:
[0065] (1) Data collection: collect target protein and its activity data.
[0066] Collect multiple protein targets from the database and literature, and then download the activity data of each protein target from the ChEMBL database. The activity data includes: IC50 (half maximal inhibitory concentration, half inhibitory concentration), which refers to the half maximal inhibitory concentration when the specified biological process is inhibited required drug or inhibitor concentration), EC50 (halfmaximal effective concentration, half maximum effect concentration), refers to the concentration of drug or inhibitor that can reach 50% of the maximum biological effect after a specific exposure time), inhibition constant (Ki value, the half-dissociation concentration of the drug) in the data of any activity value. The final activity data value in this embodiment i...
Embodiment 2
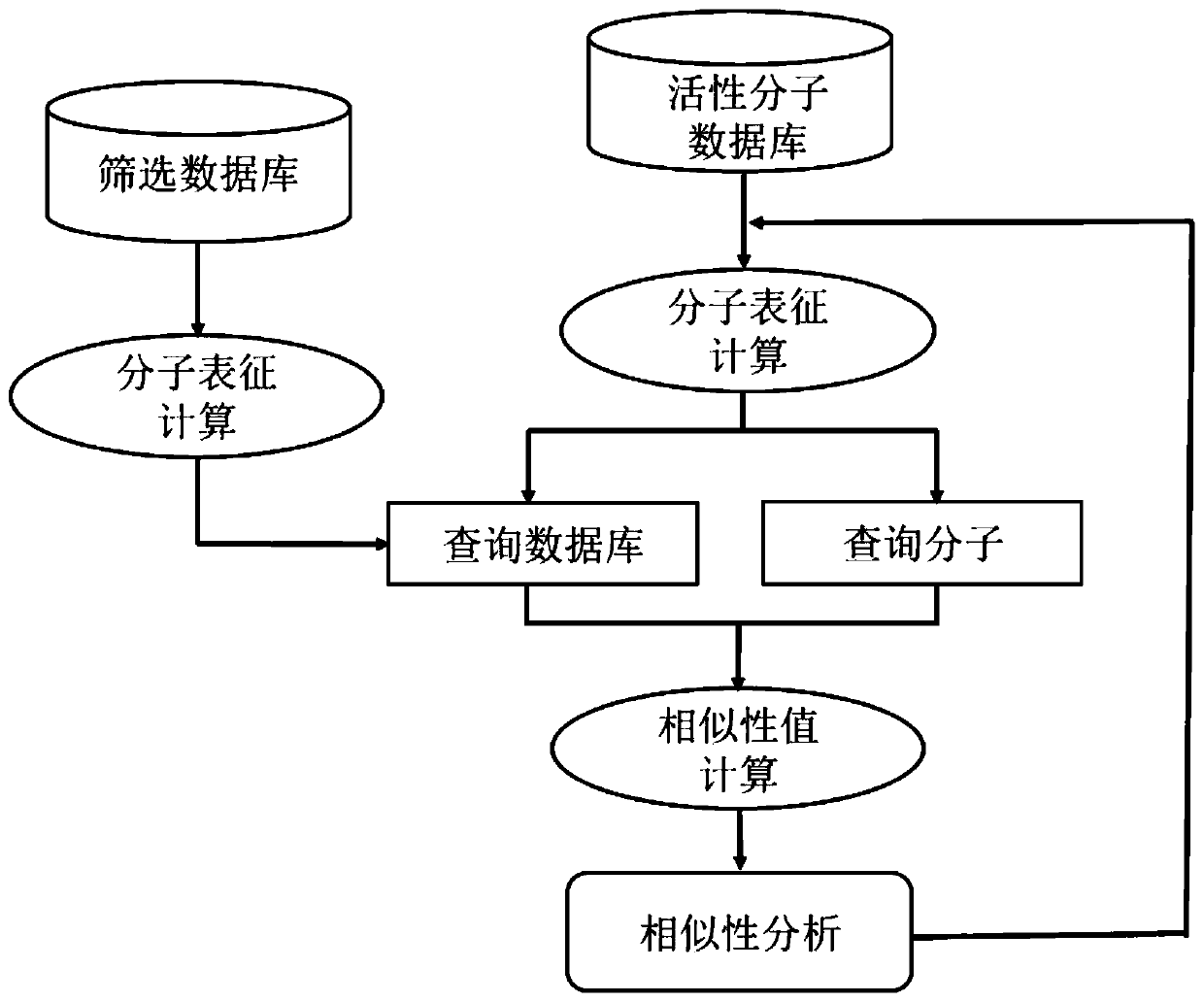
[0084] An application of the calculated target spectrum of embodiment 1 in the skeleton transition, its application process can be found in figure 2 , the specific construction steps are:
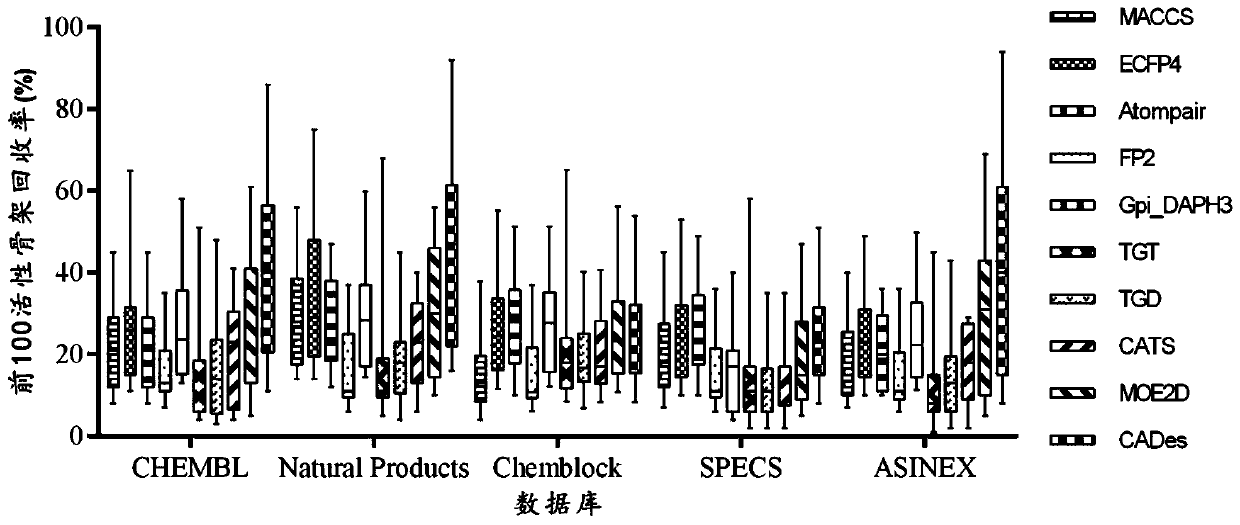
[0085] (1) Active molecule database: used from the literature: Vogt, M.; Stumpfe, D.; Geppert, H.; Bajorath, J. Scaffold Hopping Using Two-Dimensional Fingerprints: True Potential, Black Magic, or a Hopeless Endeavor? Guidelines for VirtualScreening.J Med Chem 2010,53,(15),5707-5715. The constructed database includes active molecules of 17 targets, and only select Ki or IC50 less than 1μm, the number of heavy atoms within 10-50 and side A molecule in which the number of non-hydrogen atoms in the chain group is less than the number of non-hydrogen atoms in the backbone. Each target has at least 10 different Bemis-Murcko frameworks (proposed by Bemis and Murcko, defined as a scaffold extracted from a molecule that removes the R-group but retains the linker between the ring systems), and eac...
Embodiment 3
[0094] An application of the computational target profile of Example 1 to predict protein targets, the construction process of which is as follows:
[0095] (1) The drug bromocriptine is input into the consensus model, and the calculated target spectrum of bromocriptine is output to form a molecular characterization.
[0096] (2) We will calculate the predicted probability value in the target spectrum as the binding affinity value between the compound and the protein target, and rank the targets according to the binding affinity value.
[0097] (3) Bromocriptine was shown in computational target profiling to potentially interact with a novel protein D(1A) dopamine receptor (UniprotID: Q95136). Bromocriptine mesylate is a semisynthetic ergot alkaloid derivative with strong dopaminergic activity and is indicated for the treatment of Parkinson's disease. By consulting relevant literature, it was found that the binding affinity between bromocriptine and D(1A) dopamine receptors (...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com