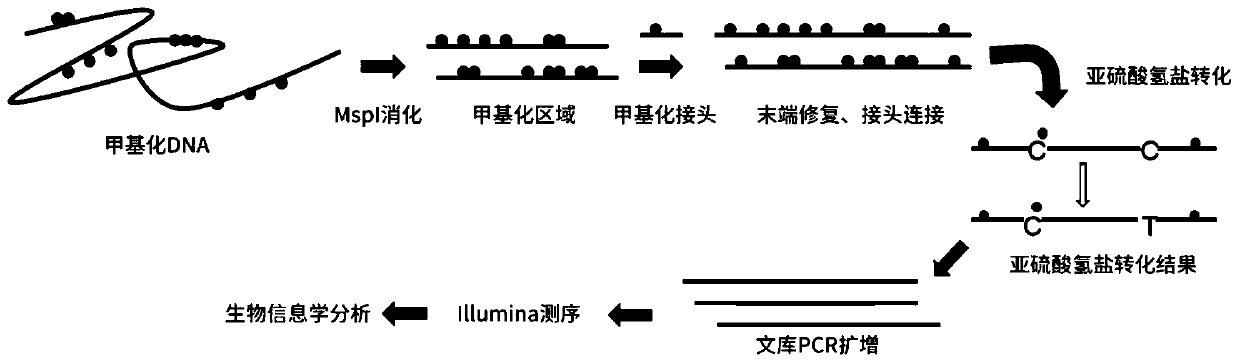
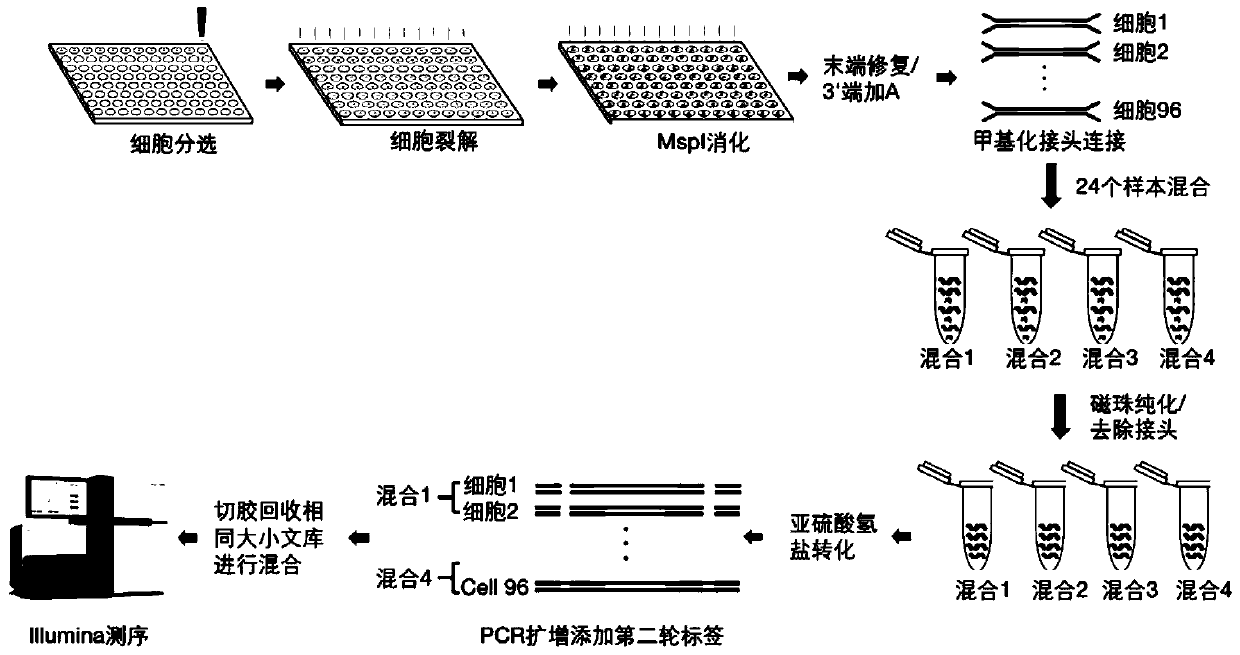
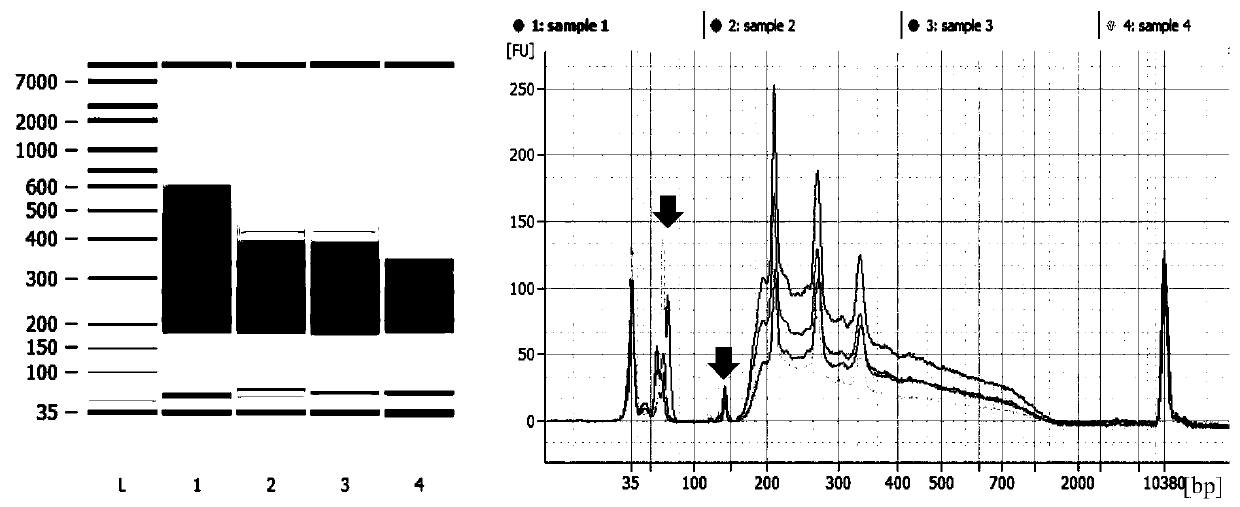
Method for constructing multiple single-cell simplified representative methylation library based on Illumina sequencing platform
A sequencing platform and construction method technology, applied in the field of molecular biology, can solve problems such as DNA loss and DNA fragmentation, and achieve the effect of improving coverage and saving construction time
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0076] Example 1 single cell lysis
[0077] 1. Dispense 5 μl of 0.1x CutSmart Buffer to each well in a 96-well PCR plate. The CutSmart buffer, that is, the cell lysate component is 10 mM Tris-HCl (PH=7.0), 10 mM EDTA, 10 mM NaCl and 0.1% (wt / vol) SDS.
[0078] 2. Sorting one single cell per well in a 96-well plate. If the sorted cells are not ready for making single-cell reduced representative methylation libraries, the 96-well plate can be sealed with a PCR Seal "F" membrane (BIORAD, CAT#MSF1001) and the It was stored in a -80°C freezer until further processing.
[0079] 3. Prepare the cell lysis buffer mix according to the table below.
[0080]
[0081] Note: Prepare an additional 20% more master mix to compensate for loss of reagents during pipetting.
[0082] 4. Distribute 19 μl of the above buffer solution to each well of 12 wells in row A of a 96-well PCR plate, seal the 96-well plate and perform short centrifugation, and then use a 12-channel pipette to transfer ...
Embodiment 2
[0090] Embodiment 2 Restriction endonuclease digestion
[0091] 1. Prepare the MspI digestion reaction mixture as follows:
[0092]
[0093] Note: Another methylation-insensitive restriction enzyme can be added to increase CpG island coverage. Make sure different restriction enzymes are compatible in the same CutSmart buffer.
[0094] 2. Dispense 19 μl of the Msp I digestion reaction mixture into each of the 12 wells in row C of a 96-well PCR plate.
[0095] 3. Carefully remove the cover film of the 96-well sample plate to avoid cross-contamination of the sample, and then use a 12-channel pipette to transfer 2 μl of the digestion mixture to each sample well. Briefly centrifuge the 96-well plate.
[0096] 4. Mix the reaction by shaking gently and centrifuge briefly.
[0097] 5. Put the sample plate into the thermal cycler, and the temperature of the thermal cover is preset to 80°C. Follow the procedure below to incubate the reaction:
[0098] program Temper...
Embodiment 3
[0101] Example 3 End Repair
[0102] 1. Prepare the end repair system according to the following formula:
[0103]
[0104] 2. Distribute 19 μl of the above reaction system to each well of 12 wells in row E of a 96-well PCR plate.
[0105] 3. Carefully remove the cover membrane of the 96-well sample plate to avoid cross-contamination of samples, and then use a 12-channel pipette to transfer 2 μl of end repair mixture to each sample well. Briefly centrifuge the 96-well plate.
[0106]4. Mix the reaction by shaking gently and centrifuge briefly.
[0107] 5. Put the sample plate into the thermal cycler, and the temperature of the thermal cover is preset to 80°C. Follow the procedure below to incubate the reaction:
[0108] program Temperature(°C) Duration (Minutes) 1 30 10 2 37 10 3 70 5 4 4 hold
[0109] 6. After the incubation, centrifuge the sample plate at 3000 rpm for 30 seconds.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com