Application of anti-BASIGIN humanized antibody in preparation of drugs for treating COVID-19
A technology of humanized antibody and coronavirus, applied in the direction of antiviral agents, antibodies, drug combinations, etc., can solve the problems of blocking antibodies, allergic and toxic reactions, etc., and achieve the effect of inhibiting chemotaxis
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
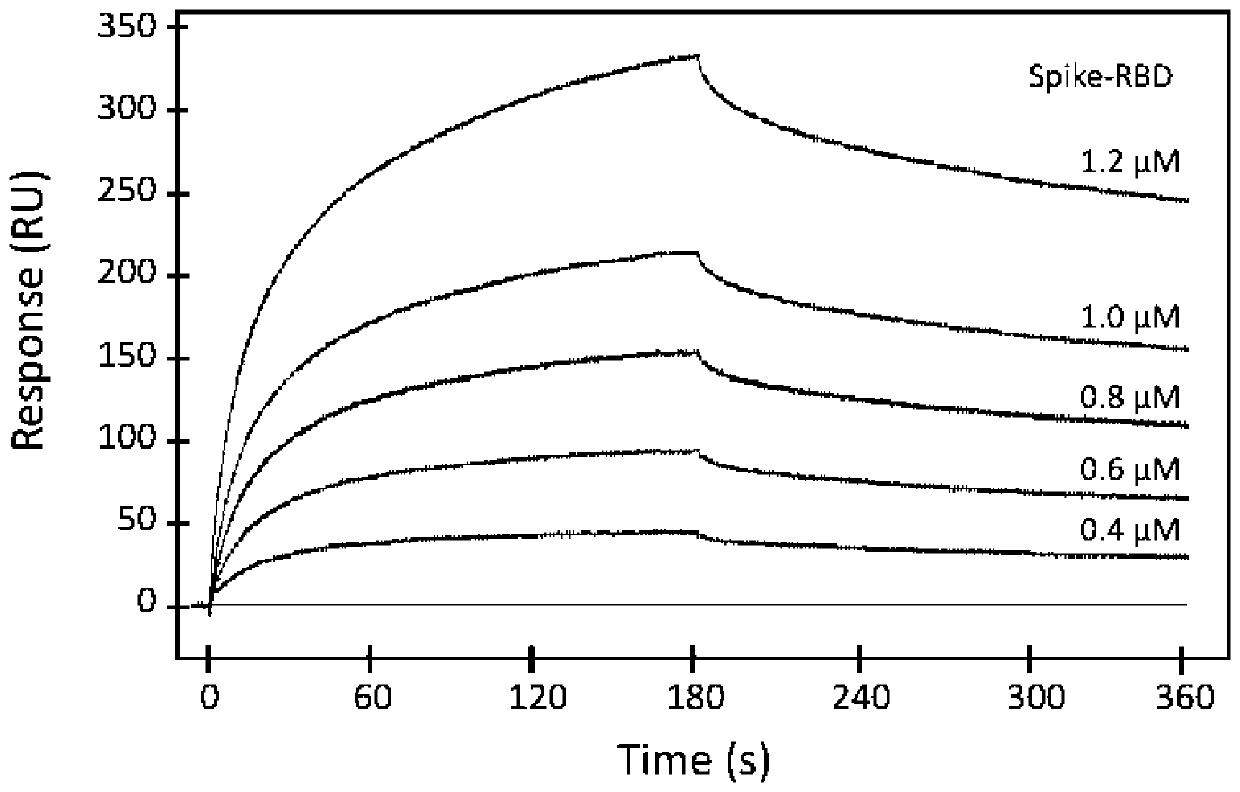
[0053] Example 1: The SPR method measures the interaction between the new coronavirus S protein (COVID-19Spike-RBD) and CD147
[0054] Based on surface plasmon resonance (SPR) technology, BIAcore 3000 system and CM5 sensor chip were used to measure the molecular binding kinetic parameters between the new coronavirus S protein (COVID-19Spike-RBD) and CD147. The detection buffer system is PBS (pH7.4), and the detection temperature is 25°C; 35 μg CD147 is immobilized on the surface of the CM5 chip by amino coupling; the interaction is detected by Kinetic Analysis / Concentration Series / Direct Binding mode, and the flow rate is set to 10 μL / min , the binding time is 10min, and the dissociation time is 10min; 0.4, 0.8, 1.0 and 1.2μM of the new coronavirus S protein (COVID-19Spike-RBD) of the samples to be tested are injected into different channels respectively. The results were fitted by BIAevaluation software to determine the kinetic constants.
[0055] The result is as figure 1S...
Embodiment 2
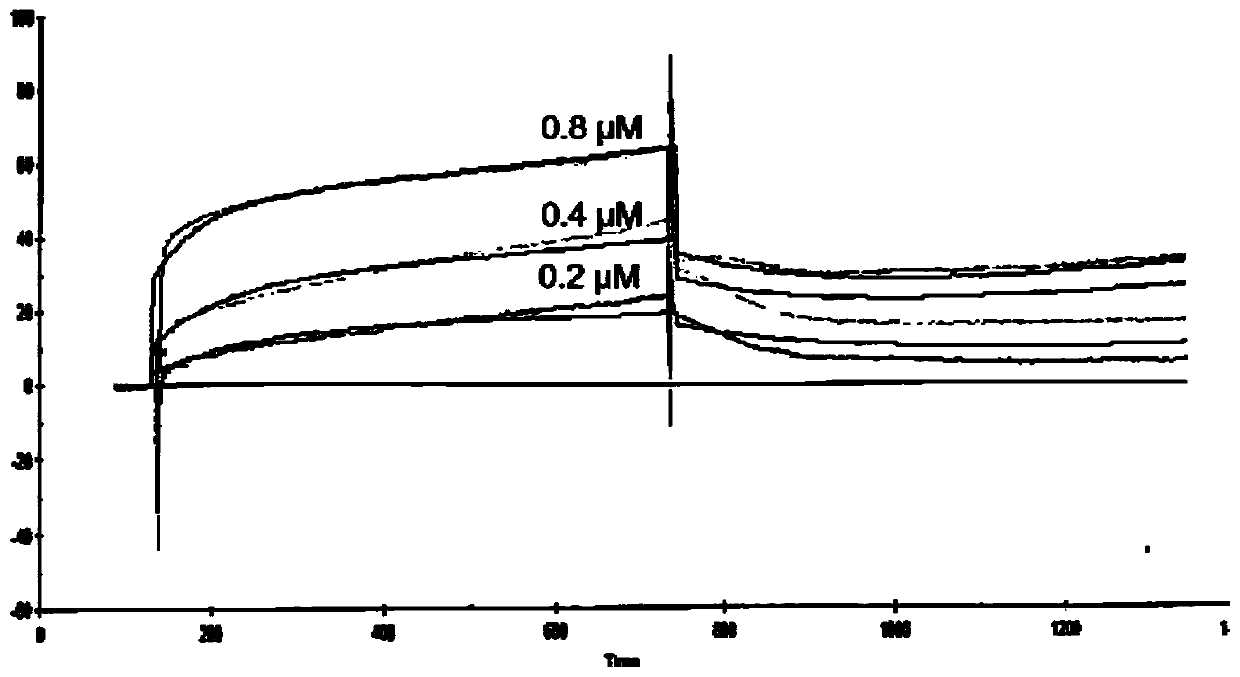
[0056] Example 2: The SPR method measures the interaction between the N protein of the new coronavirus and cyclophilin A (CyPA)
[0057] Based on surface plasmon resonance (SPR) technology, BIAcore 3000 system and CM5 sensor chip were used to determine the molecular binding kinetic parameters between the new coronavirus N protein and cyclophilin A (CyPA). The detection buffer system is PBS (pH7.4), and the detection temperature is 25°C; CyPA is immobilized on the surface of the CM5 chip by amino coupling; the chip is inactivated with 1M ethanolamine-HCl (Bio-Rad); Buffer (PBS / 0.005% Tween 20) was washed until the baseline stabilized. Inject 0.2, 0.4, and 0.8 μM of the new coronavirus N protein on the horizontal channel respectively, set the flow rate to 10 μL / min, the binding time to 10 minutes, and the dissociation time to 10 minutes; use the Kinetic Analysis / Concentration Series / Direct Binding mode to detect the interaction. The results were fitted by BIAevaluation software...
Embodiment 3
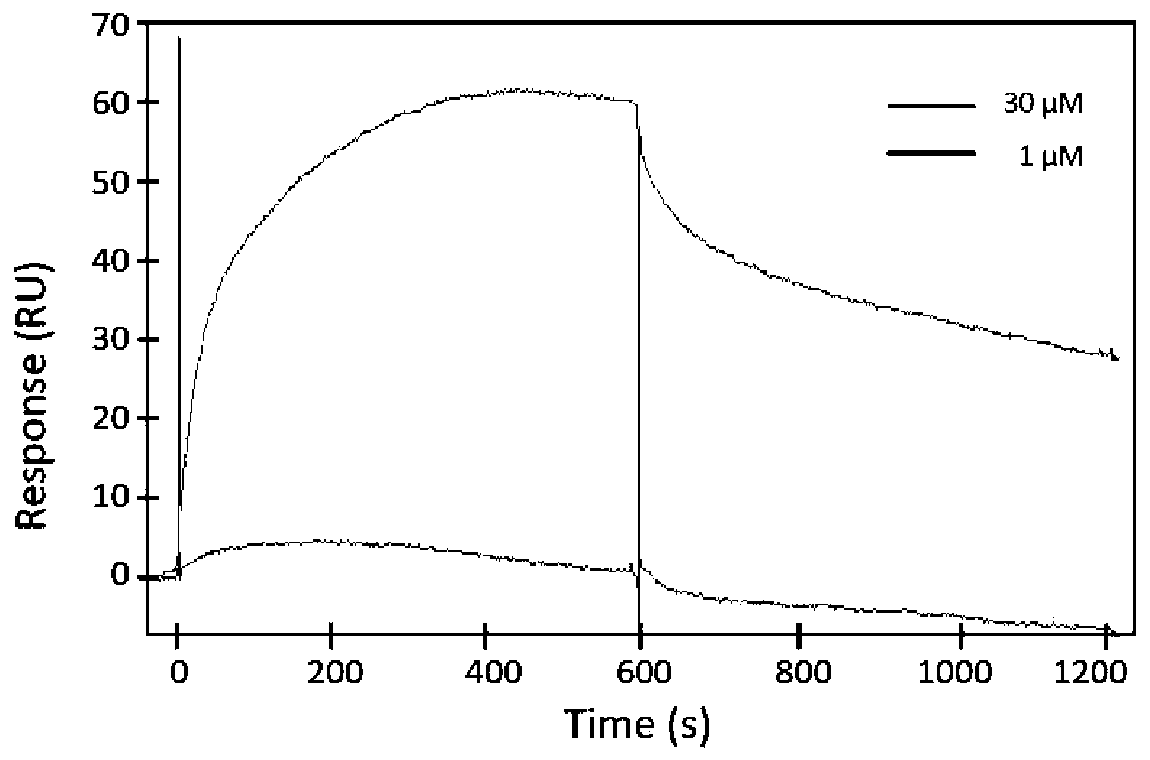
[0059] Embodiment three: SPR method measures the interaction between CD147 and cyclophilin A (CyPA)
[0060] SPR was used to monitor the binding of CD147 to cyclophilin A (CyPA) in real time, and the affinity between CD147 and CyPA was reflected by measuring the affinity constant (KD). Affinity determination of CD147 and cyclophilin A (CyPA) was performed with ProteOn XPR36 (Bio-Rad, XPR36) instrument. A GLC chip (Bio-Rad, 1765011) was activated with 0.04M EDC+0.01M sulfo-NHS (Bio-Rad). Dilute CD147 to 10mM with 10mM NaAc (pH 4.5), and inject it onto the chip at a speed of 30ul / min to couple the antigen to the activated chip through amino groups. Finally, the chip was inactivated with 1M ethanolamine-HCl (Bio-Rad); the chip was rotated 90 degrees and washed with buffer solution (PBS / 0.005% Tween 20) until the baseline was stable. 1 and 30 μM cyclophilin A (CyPA) samples were injected on the horizontal channel, respectively. The injection speed is 30ul / min. The sample bindi...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Affinity constant | aaaaa | aaaaa |
| Affinity constant | aaaaa | aaaaa |
| Affinity constant | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com