Primer set, kit and detection method for rapid detection of carbapenemase genes
A carbapenemase and primer set technology, applied in biochemical equipment and methods, recombinant DNA technology, resistance to vector-borne diseases, etc., can solve the problem of not detecting genes at the same time, and shorten the detection time and save the detection. Cost and effect of fully closed reaction process
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0048] Composition and detection method of a kit for rapidly detecting carbapenemase gene
[0049] 1. The composition of the kit
[0050] (1) Primer set: amplification primer pairs and molecular beacon probes for detecting Enterobacteriaceae carbapenemase resistance genes KPC, NDM, OXA48, VIM and IMP;
[0051] Wherein, the primer set of KPC gene is:
[0052] KPC-F (SEQ ID NO: 1): CAGCTCATTCAAGGGCTTTC;
[0053] KPC-R (SEQ ID NO: 2): CGTCATGCCTGTTGTCAGAT;
[0054] Molecular beacon probe KPC-probe (SEQ ID NO: 3): cgcgaACACACCCATCCGTTACGGCA tcgcg;
[0055] The primer set for the NDM gene is:
[0056] NDM-F (SEQ ID NO: 4): ATATCACCGTTGGGATCGAC;
[0057] NDM-R (SEQ ID NO: 5): TAGTGCTCAGTGTCGGCATC;
[0058] Molecular beacon probe NDM-probe (SEQ ID NO: 6): cgcgaGCCTGATCAAGGACAGCAAGG tcgcg;
[0059] The primer set for the OXA48 gene is:
[0060] OXA48-F (SEQ ID NO: 7): GTGGCATCGATTATCGGAAT;
[0061] OXA48-R (SEQ ID NO: 8): AGAGCACAACTACGCCCTGT;
[0062] Molecular beacon probe...
Embodiment 2
[0082] Utilize the method for rapid detection carbapenemase gene of the kit described in embodiment 1
[0083] 1. Sample
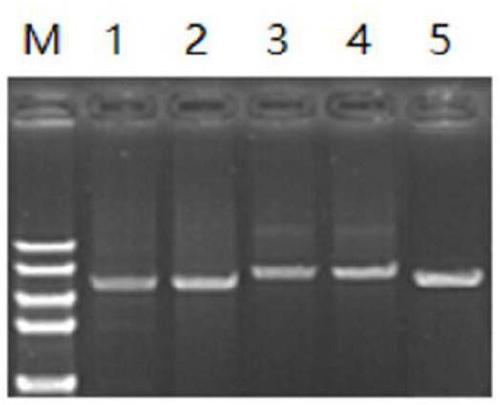
[0084] The CRE strains of ST11 Klebsiella pneumoniae isolated from clinical samples were randomly collected, and then the isolated CRE strains were cultured at 37°C for 24 hours, and then the DNA was extracted using the method described in Example 1. Five common Enterobacteriaceae carbapenemase drug-resistant gene KPC, NDM, OXA48, VIM and IMP specific primers, the corresponding positive samples were screened by PCR and identified by sequencing, and then a single carbapenemase drug-resistant gene KPC, DNA samples of NDM, OXA48, VIM and IMP were mixed to make DNA samples containing five common Enterobacteriaceae carbapenemase resistance genes KPC, NDM, OXA48, VIM and IMP. The clinical sample used in this embodiment is a blood sample, but it is not limited thereto. In addition, respiratory samples such as urine and sputum containing the CRE strain of Klebsie...
Embodiment 3
[0090] Sensitivity Analysis of Isothermal Amplification Melting Curve Detection Kit Based on Molecular Beacon Technology for CRE Strains Isolated from Clinical Samples
[0091] 1. Method
[0092] Sample processing: The CRE strain of ST11 Klebsiella pneumoniae isolated from clinical samples was adjusted to 1.8×10 with normal saline. 8 CFU / mL, then carry out gradient dilution (100 μL bacterial solution + 900 μL normal saline for dilution), then centrifuge at 10,000 rpm for a short time, discard the supernatant, then crush it on a shaker for 10 minutes, take 300 μL of the supernatant and add into a 96-deep-well plate, extract DNA on a nucleic acid extractor, and use 1.5×10 7 CFU / mL-1.5×10 2 The DNA extracted from the CFU / mL bacterial solution was used as a template, and 5 μL of each concentration gradient was amplified.
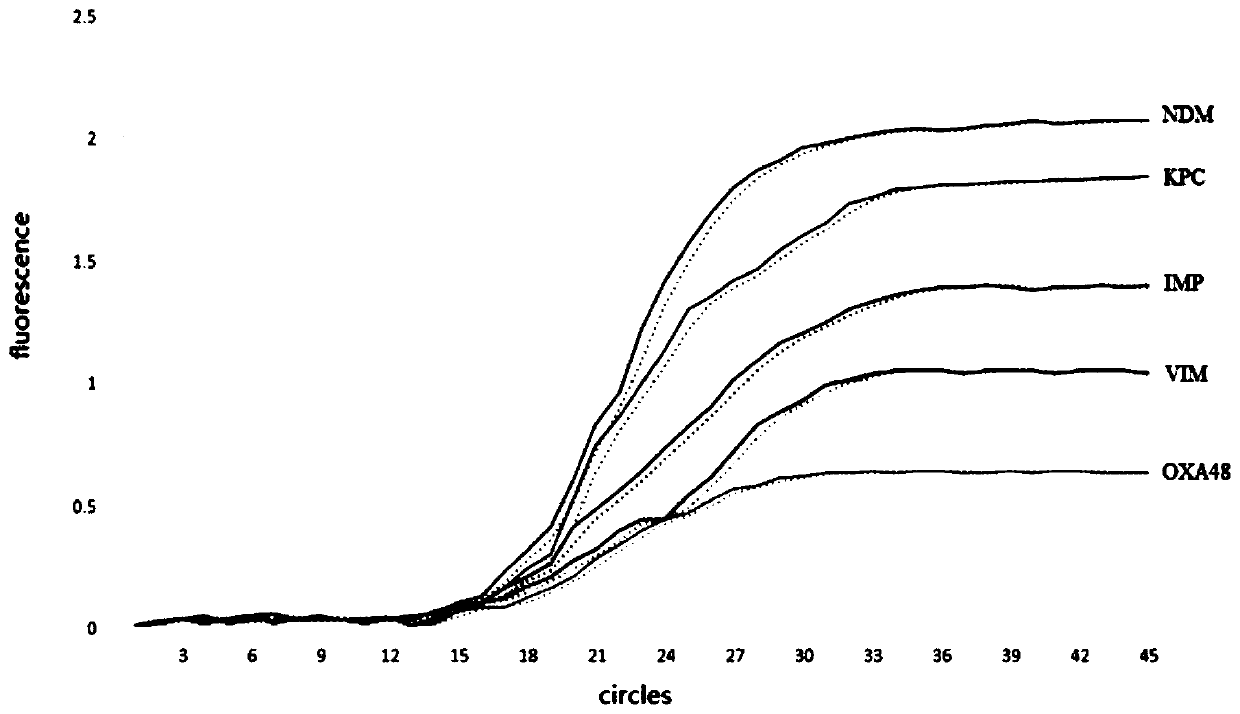
[0093] 2. Use the detection method described in Example 1 to detect. After the reaction is completed, observe the peak Tm value of the melting curve, and an...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com