Fusarium proliferatum SSR molecular marker and application thereof
A technology for the generation of Fusarium spp. and molecular markers, which is applied in the determination/inspection of DNA/RNA fragments, instruments, microorganisms, etc. Insufficient genetic variation of bacteria, etc., to achieve the effect of good repeatability and high polymorphism
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0033] Example 1: Genome-wide SSR scanning and primer design of Fusarium sp.
[0034] Considering the availability, representativeness and importance of the data, the whole genome sequence of the emerging Fusarium sp. ET1 strain (https: / / www.ncbi.nlm.nih.gov / genome / 2434) was downloaded from the NCBI database, using The SSR Hunter software searches the SSR sequence among them, selects 2-6 nucleotides as repeat motifs, and repeats at least 6 times. Using the detected SSR and its upstream and downstream 150bp flanking sequences, Primer 5 software was used to design SSR primers. The general principles of primer design are: primer length 19~25bp; GC content 40%~60%; annealing temperature Tm value 52~58°C, and the Tm value difference between upstream and downstream primers is not more than 5°C; PCR amplification length 100~300bp ; Try to avoid primer dimers, hairpin structures, mismatches, etc. The synthetic primers have specific fluorescent labels of 6-FAM (blue) or HEX (green) a...
Embodiment 2
[0035] Example 2: Screening of SSR molecular markers
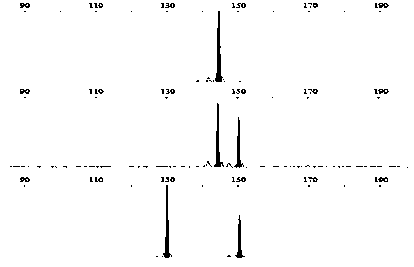
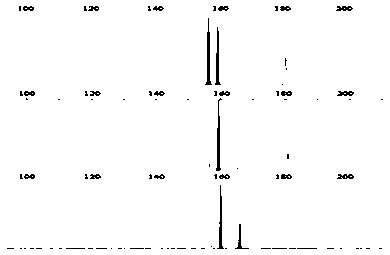
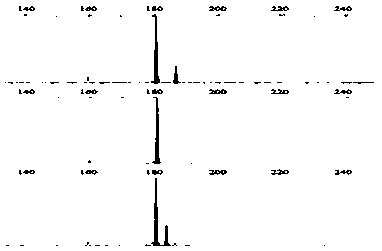
[0036] From the results of the above genome SSR scanning and primer design, 52 pairs of SSR primers were selected at random, and the specificity of these primers in the genome was detected by local Blast. After the pre-experiment, 12 microsatellite loci with diversity in the fragment length of the amplified product ranging from 100 to 300 bp were selected, and contained different simple sequence repeating units (Table 1). figure 1 , figure 2 , image 3 , Figure 4 , Figure 5 , Figure 6 , Figure 7 , Figure 8 , Figure 9 , Figure 10 , Figure 11 and Figure 12 The SSR genotypes of Fusarium sp. FP03, FP08, FP13, FP21, FP24, FP26, FP31, FP32, FP35, FP43, FP48 and FP51 are shown, respectively. The primers of the 12 SSR loci showed polymorphism, which provided markers for the analysis of the genetic diversity of Fusarium sp.
[0037] Table 1. 12 SSR molecular markers of Fusarium
[0038]
Embodiment 3
[0039] Example 3: Application of SSR Molecular Markers in Genetic Diversity of Fusarium sp.
[0040] Step 1: Collection, isolation and identification of layered Fusarium
[0041] In this example, Fusarium sp. collected from 3 populations (FY03, FY08, FY13) in Hangzhou, Zhejiang and 2 populations (NN05, NN12) in Nanning, Guangxi were used as samples. Fusarium was isolated from a susceptible sample of rice ear rot. Specific primers were designed according to the conserved sequence of the protein translation elongation factor (EF) gene, EF1 (5'-ATGGGTAAGGAGGACAAGAC-3') and EF2 (5'-GGAAGTACCAGTGATCATGTT-3'), and the strain was amplified by PCR. After the PCR amplification products were sequenced, the sequences were submitted to the Fusarium TEF sequence database (FUSARIUM-ID v 1.0) for comparison, and finally Fusarium sp. was selected by sequence comparison.
[0042] Step 2: Extraction of Genomic DNA from the Outer Fusarium Strains
[0043] The Fusarium genomic DNA was extracte...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com