Preparation method of capture probe for targeted sequencing of novel coronavirus virus SARS-CoV-2 genome
A technology for targeted sequencing and capture probes, used in microorganism-based methods, biochemical equipment and methods, and microbial determination/inspection, etc. Infection, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0025] The method of the present invention for preparing capture probes for targeted sequencing of the new coronavirus SARS-CoV-2 genome and the method for preparing capture probes for targeted sequencing of the new coronavirus SARS-CoV-2 genome of the present invention includes the following steps:
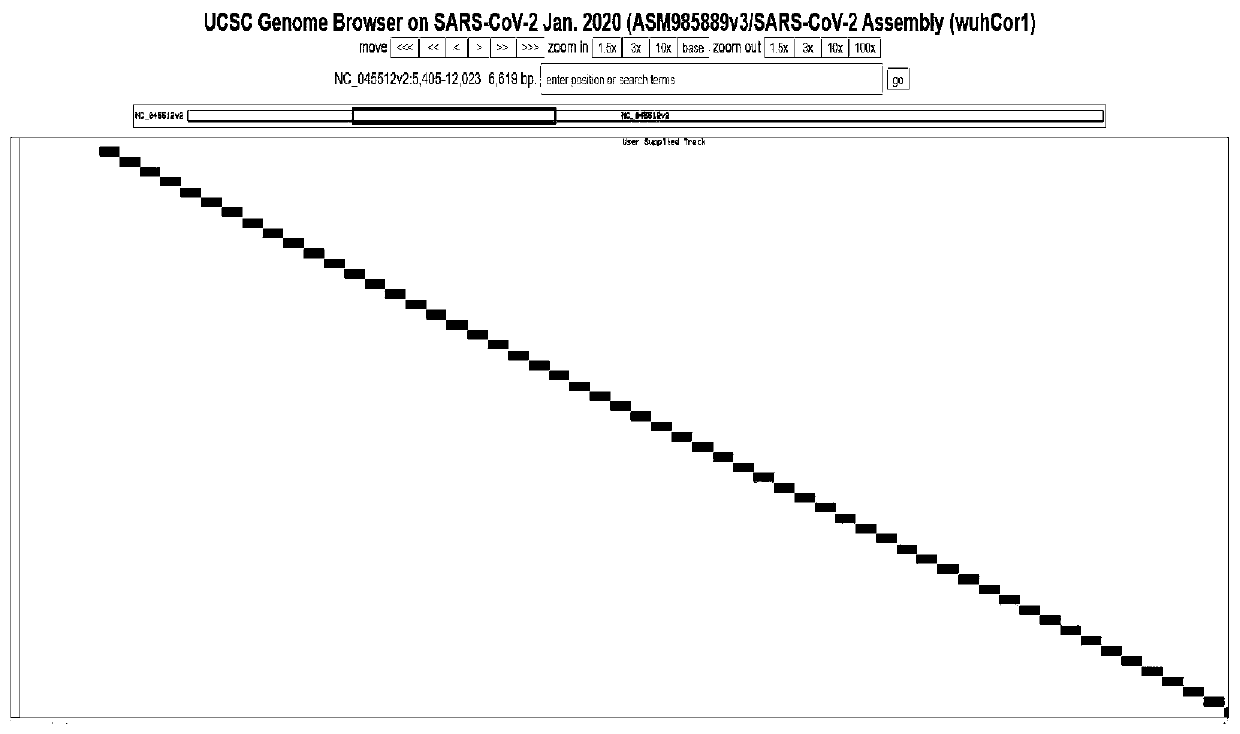
[0026] Step one, download the SARS-CoV-2 virus reference genome sequence NC_045512.2 from the NCBI database, and adopt the shingled coverage design strategy, that is, cover the probe from beginning to end, and adjust the length of the probe to 75-120nt according to the GC content;
[0027] Step two, quantitatively evaluate the probe library, and filter out unqualified probe sequences; after removing unqualified probes, 249 probe libraries are left, covering 99.23% of the entire genome of the new coronavirus;
[0028] Step 3: Prepare 249 probe library DNA templates, PCR amplification, and purification of PCR products;
[0029] Step 4: Digest the purified PCR product and smooth the ends wit...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com