Primer combination used for detecting SARS-CoV-2 whole genome, and application method
A technology of whole genome and primer combination, which is applied in the field of primer set for detecting the whole genome of SARS-CoV-2, can solve the problems of not meeting the requirements of in-depth analysis data, relying on electrophoretic separation technology, and low throughput, so as to reduce the cost of detection, High amplification efficiency and high throughput
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0059] Example 1: Primer Design and Optimization
[0060] 1. Design of primers
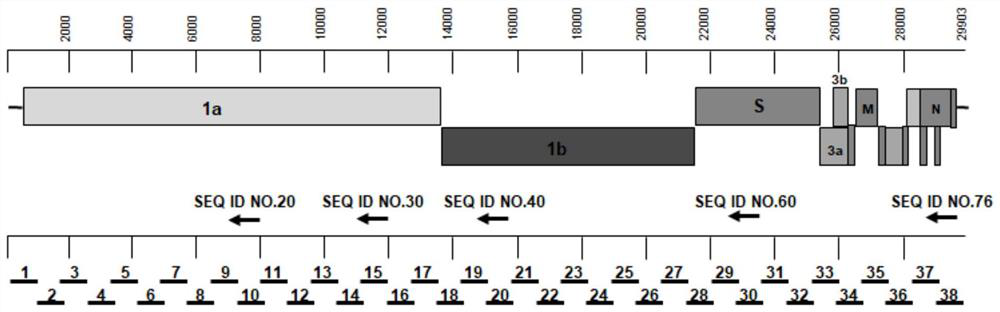
[0061] A. Due to the highly conservative nature of the novel coronavirus, download five G-type, S-type, and V-type sequences from the website of the Global Initiative for Sharing Influenza Data (GISAID) as reference sequences, compare them with the clustalx software, and select the conserved region sequences , the GC content, Tm value, hairpin structure and primer-dimer were analyzed by Primer Select software. Only when all qualified can indicate that the preliminary design of the primer is completed, the full-length sequence gene structure map of SARS-CoV-2 and 5 specific reverse transcription Primer positions and 38 sets of primers to amplify segments such as figure 1 shown;
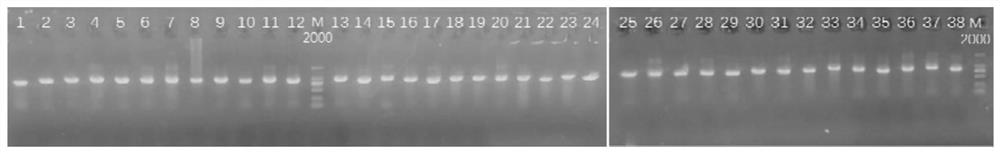
[0062] B. There is an overlapping region of 100-200bp in every two fragments of the 38 fragments to facilitate splicing and complete splicing. The primer sequences are shown in SEQ ID NO:1-SEQ ID NO:152, see the table b...
Embodiment 2
[0068] Embodiment 2: the extraction of sample RNA
[0069] In this example, the commercialized TIANamp Virus RNA Kit (TIANGEN, Beijing) was used to extract RNA from throat swabs or nasal swabs of 20 patients with new coronary pneumonia. The extraction method refers to the kit instructions.
Embodiment 3
[0070] Embodiment 3: reverse transcription nested PCR reaction
[0071] 1. Reverse transcription
[0072] Carry out RT-PCR with the RNA gained in embodiment 2 as template, and reaction is carried out in two steps, and reaction system and reaction condition are as follows: A, reaction system are as follows:
[0073] The first step reaction system is 20 μL:
[0074]
[0075] The second step reaction system is 40 μL:
[0076]
[0077] B. The reaction conditions are as follows:
[0078] Step 1: 65°C, 5min
[0079] The second step: 47°C, 80min; 70°C, 15min.
[0080] 2. Nested PCR reaction
[0081] The reverse transcription reaction product cDNA obtained in step 1 is diluted one-fold and used as a template for a nested PCR reaction. The reaction system and conditions are as follows, and the nested PCR is carried out in two steps:
[0082] (1) Peripheral PCR:
[0083] The reaction system is 20 μL:
[0084] Reagent components Premix Taq dd H 2 o
upstre...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com