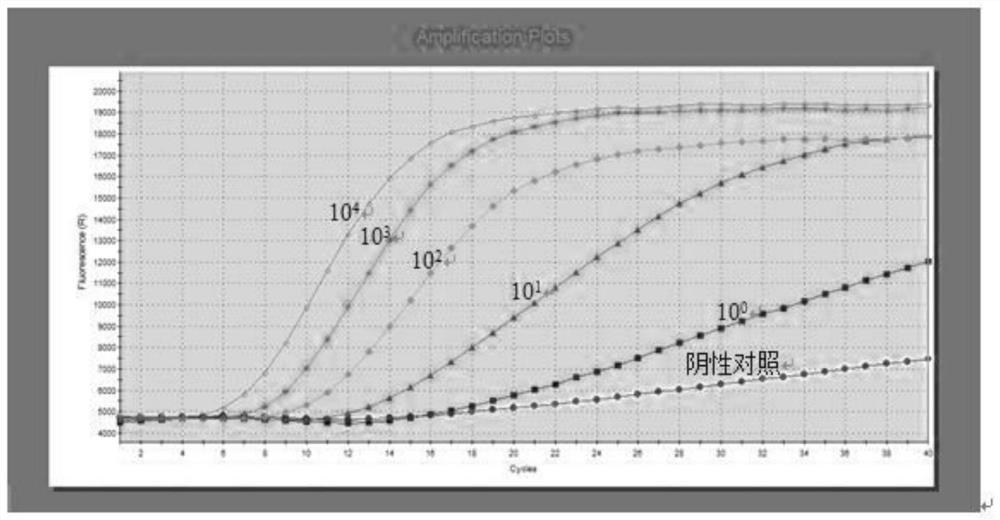
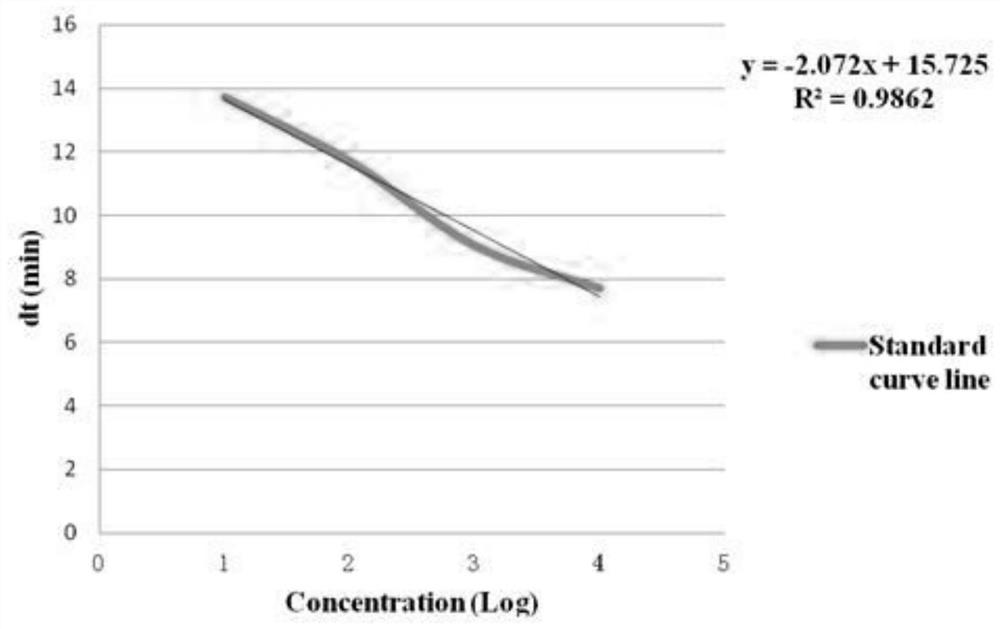
Escherichia coli nucleic acid assay method using RNA constant temperature amplification
A constant temperature amplification technology for Escherichia coli, applied in the field of environmental specimen detection, can solve the problems of time-consuming, labor-intensive, detection impact, complicated operation process, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0016] Embodiment 1: the development of Escherichia coli (Ec) nucleic acid detection kit (RNA constant temperature amplification) detection reagent
[0017] 1. Design of capture probes, primers and target nucleic acid detection probes: by comparing and analyzing the existing Escherichia coli nucleic acid sequences in the Genbank database and the nucleic acid sequences reported in published literature at home and abroad, the 16SrRNA of Escherichia coli Conserved gene fragments are the target sites for amplification, and primers and probes are designed for highly conserved segments with no secondary structure.
[0018] 2. Design of internal standard detection probe: According to the principle that the competitive internal standard probe should have the same GC content and similar Tm as the detection probe, design an internal standard probe that is completely different from the target detection probe sequence and has a different fluorescent label. Needle.
[0019] 3. Constructio...
Embodiment 2
[0035] Embodiment 2: Escherichia coli (Ec) nucleic acid detection kit (RNA constant temperature amplification) and its use
[0036] 1. Take 1ml of seawater sample into a sterile sample tube, add an equal volume of sample preservation solution, vortex and oscillate for 1min, make it fully mixed, and store for later use.
[0037] 2. Prepare the sample processing tube (1.5ml centrifuge tube, the quantity is: the number of samples to be tested + 5), and mark the number of the sample to be processed and the positive control and negative control respectively;
[0038] 3. Divide the sample processing tubes in step 2 above into 3 groups, which are sample group, positive standard group and negative control group. Add 200ul of nucleic acid extraction solution to each treatment tube; add 250ul of positive standards of different concentrations to the positive standard group; continue to add 250ul of seawater samples to the sample group, and continue to add 250ul of negative standard to th...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com