Fourfold real-time fluorescent PCR identifying and detecting method for porcine circoviruses 1 to 4
A technology of porcine circovirus and real-time fluorescence, which is applied in the biological field to achieve the effect of improving detection efficiency, high sensitivity and good repeatability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
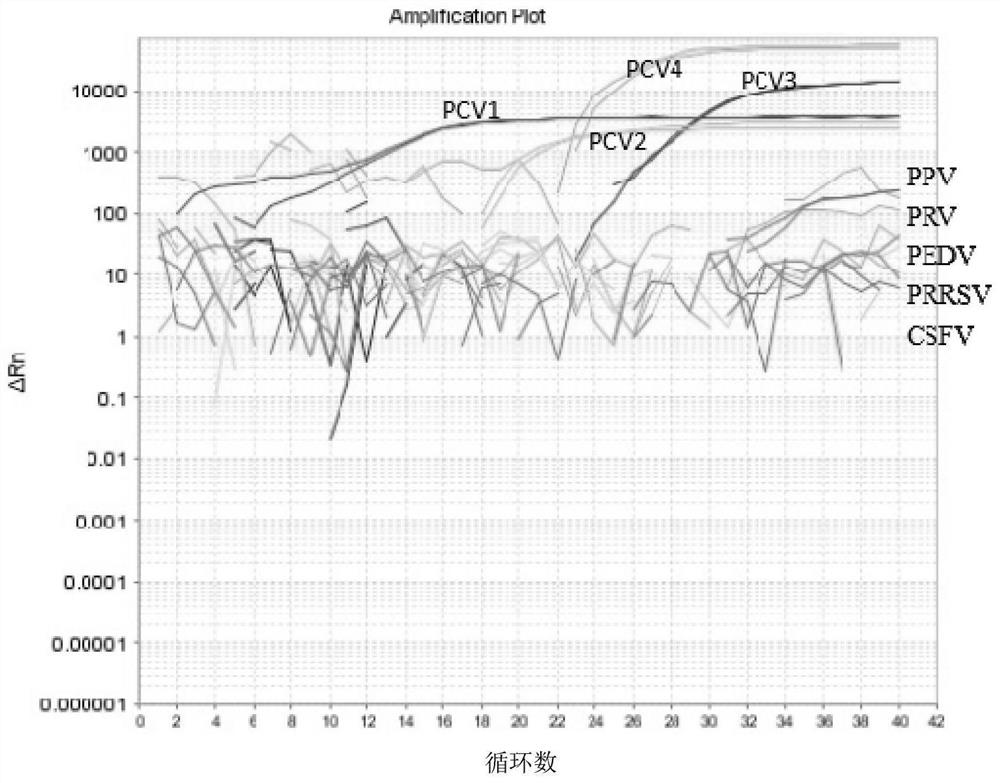
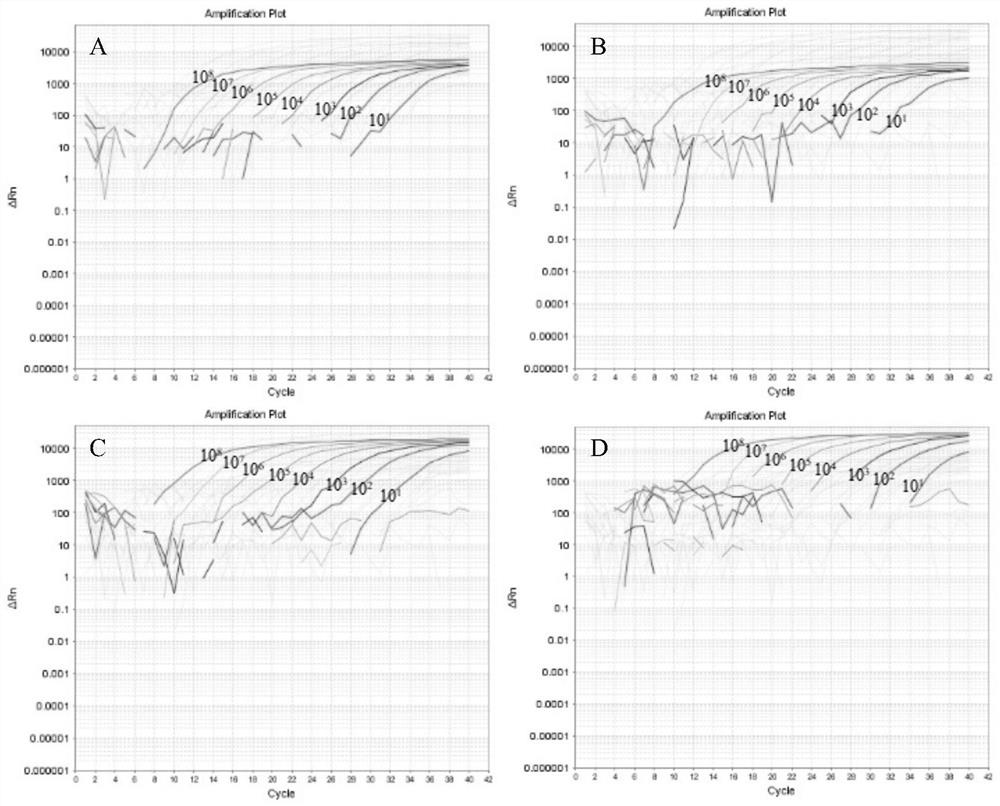
[0036] Establishment and evaluation of quadruple real-time fluorescent PCR differential detection method for porcine circovirus types 1 to 4 (PCV1-4)
[0037] 1.1 Design of primers and probes
[0038] According to the whole genome of PCV strains obtained from the GenBank database, the sequence alignment software DNAMAN was used for multiple sequence alignment analysis, and four specific TaqMans for identifying PCV1, PCV2, PCV3, and PCV4 were designed based on the highly conserved regions in the ORF1 and ORF2 genes. probe and 4 pairs of primers. The 5' ends of the probes were labeled with different fluorophores (FAM, HEX, ROX and TAMRA). The primers and probes in PCV quadruple real-time fluorescent PCR are shown in Table 1-1.
[0039] Table 1-1. Primers and probes used to construct PCV quadruple real-time fluorescent PCR differential detection method
[0040]
[0041]
[0042] *The positions of corresponding primers and probes in the genome are determined according to PC...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com