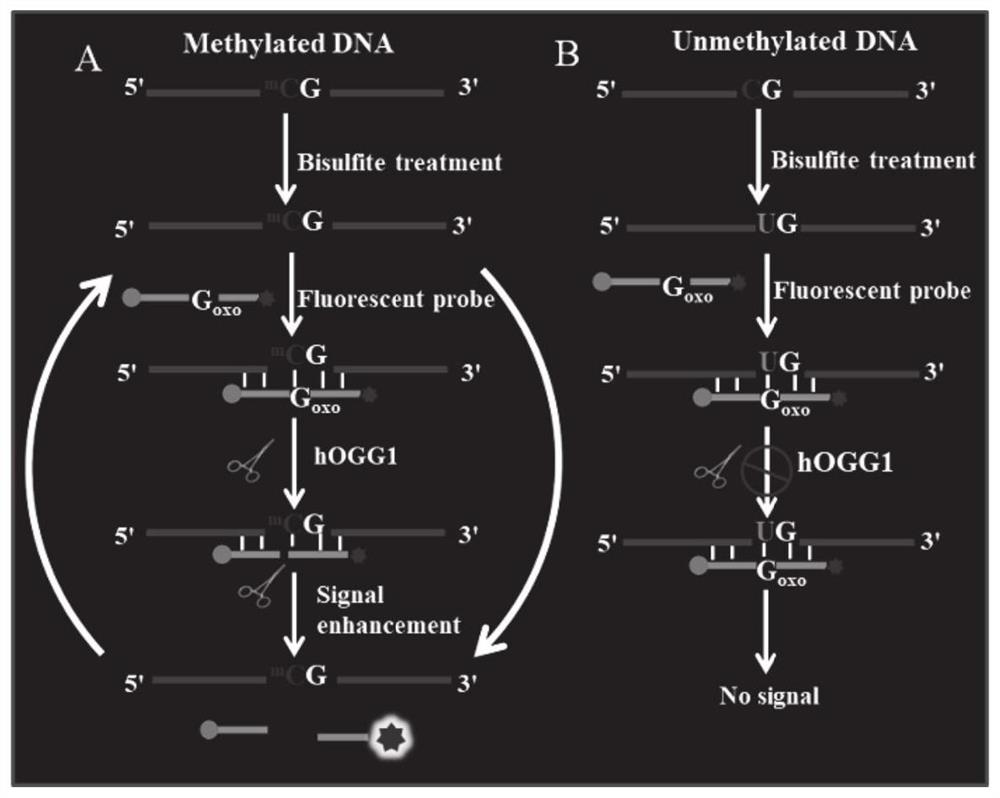
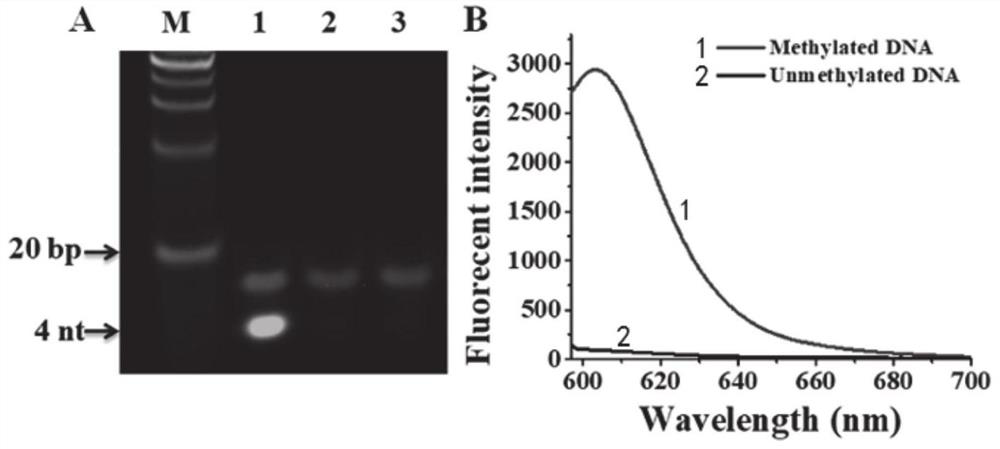
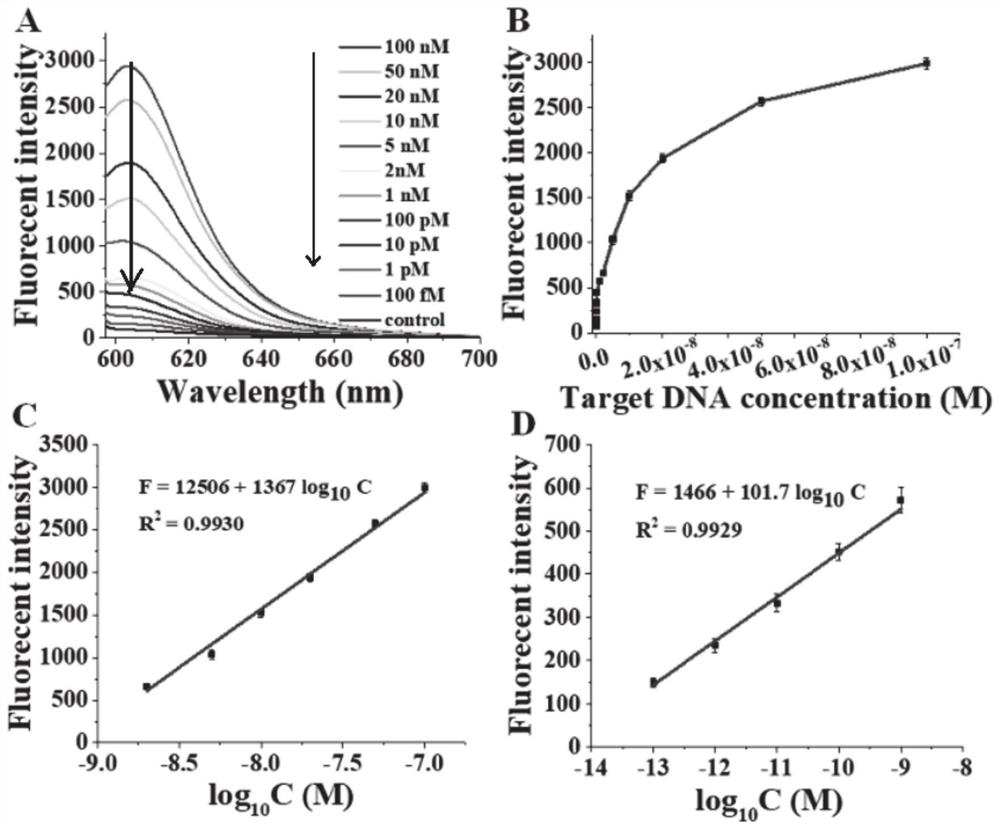
Fluorescent probe based on oxidative damage bases, and test kit and method for directly detecting DNA methylation
A fluorescent probe and oxidative damage technology, applied in the field of biological analysis, to achieve the effect of simplifying experimental procedures, high sensitivity, and improving detection sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0056] Cell culture and preparation of genomic DNA samples: Human colon cancer cells (SW480 cells) were cultured in Dulbecco's modified Eagle's medium DMEM containing 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin at 37 ℃ with 5% CO 2 cultivated in a humid environment. Genomic DNA was extracted according to the instructions of the QIAamp DNA mini kit (QIAGEN). First, cells were thoroughly lysed with lysis buffer containing proteinase K, incubated at 56°C for 10 min, and then an equal volume of 100% ethanol was added. Genomic DNA is bound to the QIAamp column, washed twice and then eluted to collect pure genomic DNA. Genomic DNA concentration was measured with a NanoDrop 2000 spectrophotometer. According to the instructions of the EZ-DNA methylation-gold kit kit, 500pg-2μg of genomic DNA was treated with bisulfite.
[0057] Bisulfite treatment of DNA: The bisulfite treatment of DNA was carried out according to the instructions of the EZ-DNA methylation-gold kit...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com