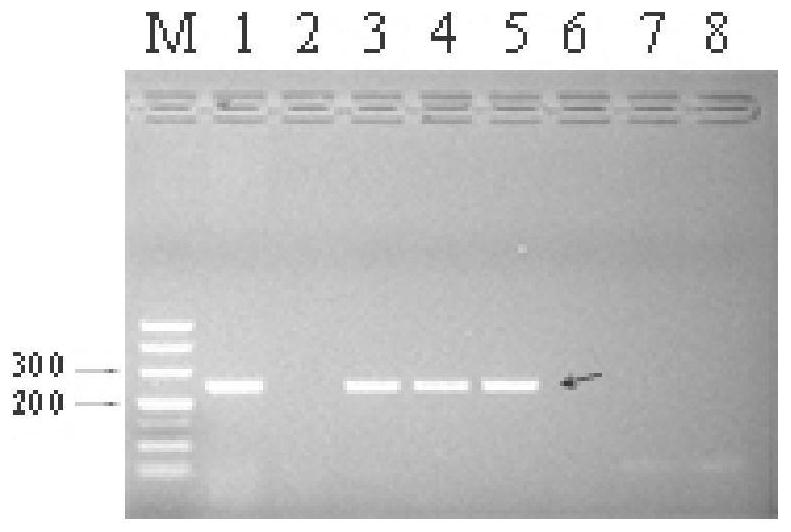
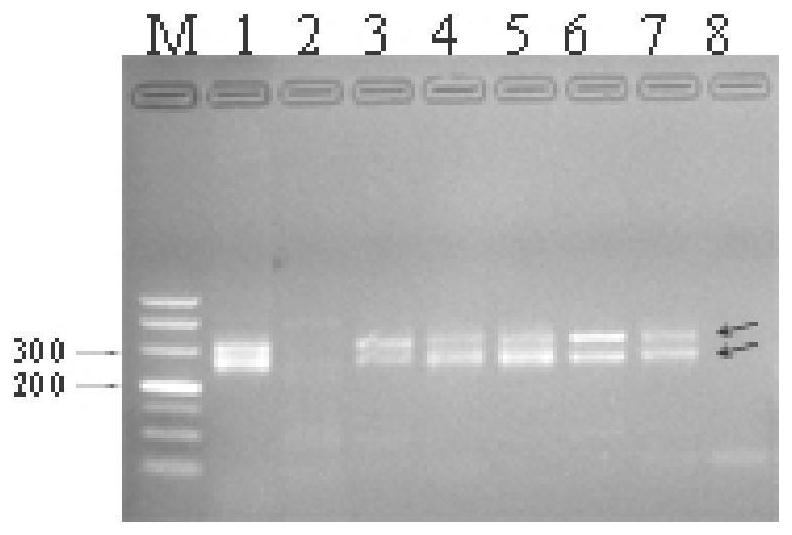
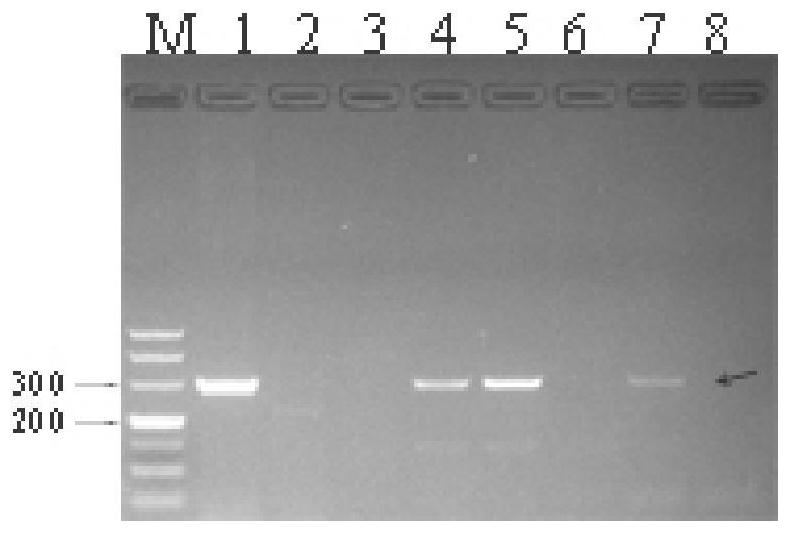
SSR molecular marker and method for identifying authenticity of filial generation of sugarcane and saccharum arundinaceum
A molecular marker and sugarcane technology, applied in the field of genetic breeding, can solve the problems of insufficient marker genome coverage, chromosome loss, and inability to effectively detect hybrid offspring, etc., to achieve clear amplification bands, speed up the breeding process, and fast, accurate and authentic Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0090] This embodiment has selected 5 different DNA mixtures of the original species of Banana (Jiangxi 83-4, Sichuan 79-I-9, Hainan 92-77, Guizhou 78-II-14), 3 sugarcane parent material DNA mixtures (tropical species Badila .
[0091] Step 1, first extract the genomic DNA of the material, and use it as a template for standby;
[0092] (1) Extraction of genomic DNA
[0093] Take the CTAB extraction method:
[0094] 1) Take the young leaves of the material, grind them with liquid nitrogen, add about 200mg of tissue samples into a 1.5mL centrifuge tube;
[0095] 2) Add 700 μL of lysis buffer preheated at 65°C, bathe in water at 65°C for 30 minutes, and turn over evenly 2-3 times during the period;
[0096] 3) Add 700 μL of chloroform / isoamyl alcohol (chloroform: isoamyl alcohol: 24:1) mixture, shake vigorously for 10 minutes, then centrifuge at 11000 rpm and 15°C for 10 minutes, and transfer the supernatant into a 1.5 mL centrifuge tube;
[0097] 4) Add an equal volume of pr...
Embodiment 2
[0114] Example 2 Identification of Pseudomonas ancestry in different backcross generations of sugarcane and Pseudomonas hybrids
[0115] The hybrid combination of 3 sugarcane cultivars (Xintaitang 22, Yuetang 00-236, Yuetang 91-976) and Banmao high-generation backcross materials (Yacheng 07-71, Yacheng 06-140) was selected:
[0116] Hybrid combination 1: Xintai Sugar No. 22 × Yacheng 07-71;
[0117] Hybrid combination 2: Yuetang 00-236×Yacheng 07-71;
[0118] Hybrid combination 3: Yuetang 91-976×Yacheng 06-140.
[0119] Seedlings were sown in the 3 hybrid combinations according to conventional methods, and sugarcane seedlings were sown in a greenhouse seedling box in sunny weather. Seedling-growing substrate, then sprinkled with sterilized sand; spray the seedling-raising substrate with water until the whole seedling-raising substrate is soaked, and finally cover with plastic film;
[0120] The seedling-raising substrate is prepared through the following steps: soil and bag...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com