Method for improving production capacity of L-histidine producing bacteria
A technology of production capacity and histidine, applied in the field of genetic engineering, can solve the problem that the production of microbial fermentation method has not yet been realized.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0048] Embodiment 1: Construction of wild-type hisG genetically engineered bacteria
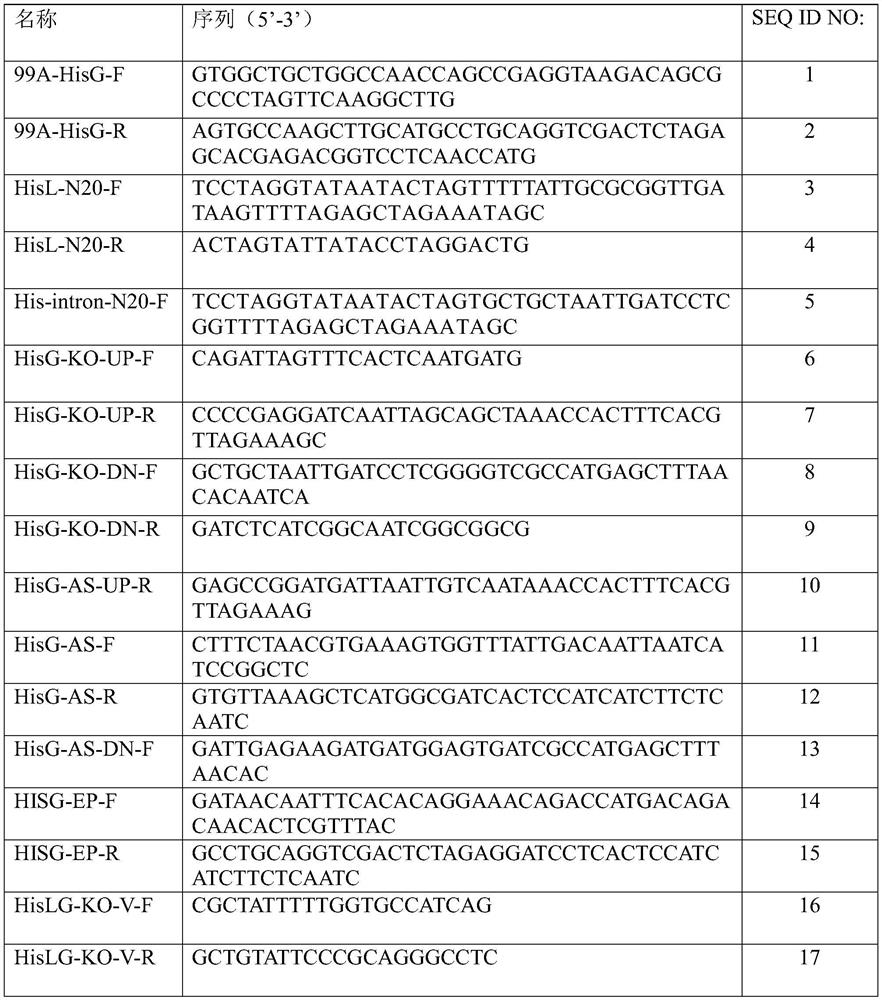
[0049] Synthetic primer pair 99A-HisG-F and 99A-HisG-R sequences SEQ ID NO: 1 and SEQ ID NO: 2, using the Escherichia coli W3110 strain genome as a template for PCR amplification (the following PCR reagents were purchased from KOD of Toyobo TOYOBO FX series).
[0050] The 50 μL PCR amplification system is: KOD FX 1 μL, KOD FX buffer 25 μL, dNTP 3 μL, genome template 0.5 μL, upstream and downstream primers 2 μL, ddH 2 O to make up 50 μL.
[0051] The PCR program was: 105°C hot lid, 95°C pre-denaturation for 5 min; 98°C denaturation for 30 s, 57°C annealing for 30 s, 68°C extension for 60 s, 30 cycles; finally 68°C extension for 10 min; 16°C cooling for 10 min.
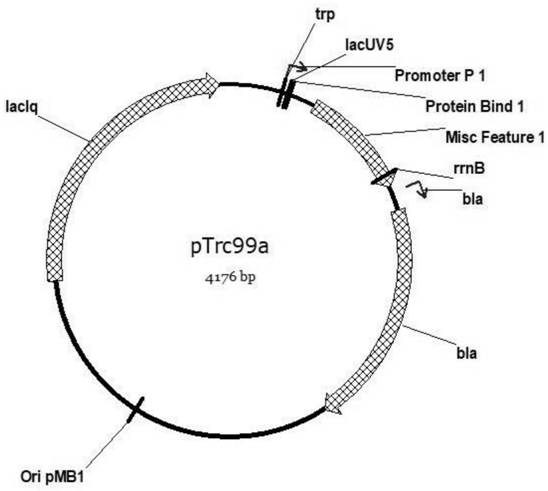
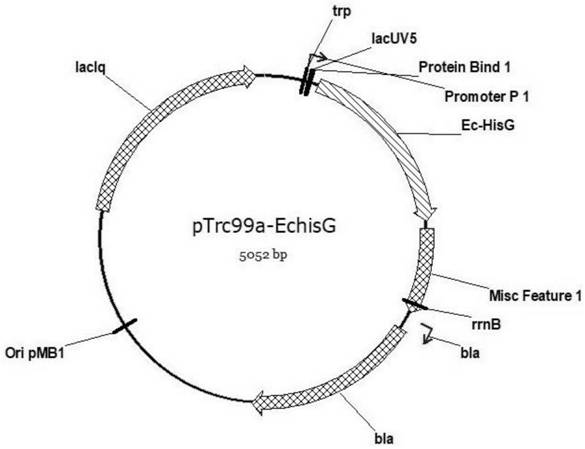
[0052] The gene SEQ ID NO: 19 of the wild-type hisG was obtained by PCR amplification. The pTrc99a plasmid was double-digested with restriction endonucleases NcoI and BamHI (see figure 1 , the plasmid was donated by researcher Yang...
Embodiment 2
[0053] Example 2: Construction of hisG random mutation point library by error-prone PCR method
[0054] Mutation of wild-type ATP phosphoribosyltransferase was performed by error-prone PCR.
[0055] The 50 μL error-prone PCR reaction system includes: 50ng of the pTrc99a-hisG plasmid template constructed in Example 1, 30pmol HISG-EP-F and HISG-EP-R primer pair SEQ ID NO:14 and SEQ ID NO:15, 1×Taq buffer, 0.2mM dGTP, 0.2mM dATP, 1mM dCTP, 1mM dTTP, 7mM MgCl2, (0mM, 0.05mM, 0.1mM, 0.15mM, 0.2mM) MnCl 2 , 2.5 units of Taq enzyme (fermentas).
[0056] The PCR reaction conditions were: 95°C for 5 minutes; 30 cycles of 94°C for 30s, 55°C for 30s, 72°C for 2min / kbp; 72°C for 10min.
[0057] Restriction endonuclease DpnI was digested at 50°C for 1 hour, and the 0.9kb mutant fragment was recovered from the gel, and the pTrc99a linearized vector constructed in Example 1 was digested with NcoI and BamHI for homologous recombination, and then transformed into the large intestine by the c...
Embodiment 3
[0058] Embodiment 3: High-throughput screening hisG mutant library
[0059] 3.1 Select the transformant in the mutant library, inoculate it into a 96-well deep-well culture plate containing 700 μL LB medium, the medium contains 100 μg / mL ampicillin, culture at 37°C for 6 hours, and add a final concentration of 0.1mM IPTG , cooled to 25°C, and incubated overnight. Centrifuge at 5000rpm for 10min, discard the supernatant, freeze at -70°C for 1h, and thaw at room temperature for 30min. Add 200 μL of 0.1M potassium phosphate buffer (pH 8.0) to resuspend the bacteria for the determination of hisG enzyme activity.
[0060] 3.2 Determination of HisG enzyme activity
[0061] For the determination of HisG enzyme activity, refer to the method provided by (Biochimie, 94, 2012, P829-838). Enzyme activity assay system: the reaction mixture contains 100mM Tris-HCl (pH 8.5), 150mM KCl, 10mM MgCl 2 , 5mM ATP, 0.5mM RPP, 1U yeast pyrophosphatase, the volume is 0.1mL. Performed in 96-well ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com