Preparation method of high-throughput sequencing library for miRNA
A sequencing library and high-throughput technology, applied in the field of high-throughput sequencing library preparation, can solve problems such as difficulty in satisfying accurate quantitative analysis, lack of accurate quantitative analysis of sequencing libraries, and inability to directly use library construction and quantitative analysis.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
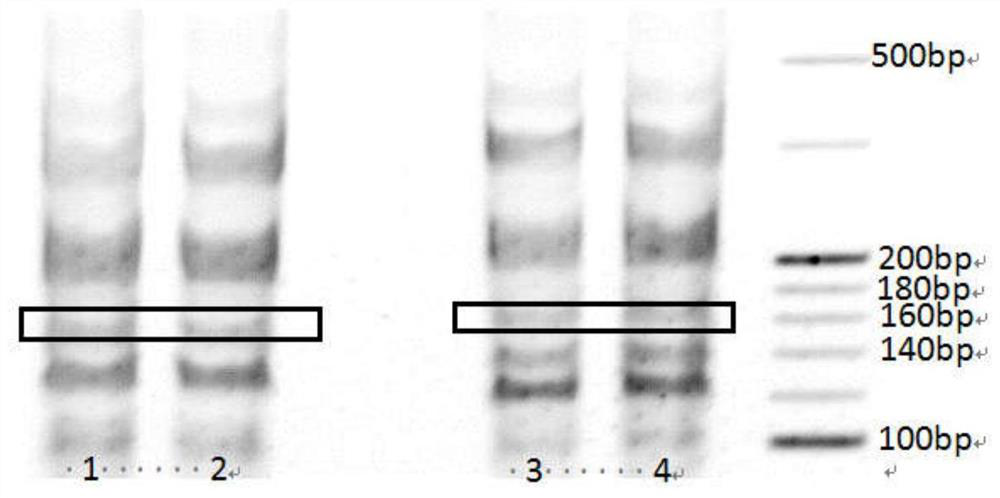
Image
Examples
Embodiment 1
[0027] 3' end plus connector
[0028] 1. Dilute a total of 10 ng of total RNA extracted from HeLa cells with ultrapure water (no DNase and RNase, the same below) to a total volume of 5 μl, and place it in a 200 μl thin-walled PCR tube;
[0029] 2. Add 1 μl adapter RA3, mix well, incubate at 70°C for 2 minutes, and immediately place on ice to cool;
[0030] 3. Add 2 μl HML (Ligation Buffer), 1 μl RNase Inhibitor, 1 μl T4RNA Ligase, mix well, and incubate at 28°C for 1 hour;
[0031] 4. Add 1 μl of STP (Stopoligo solution with a concentration of 10 mM) and mix well, incubate at 28°C for 15 minutes; add a linker at the 5' end
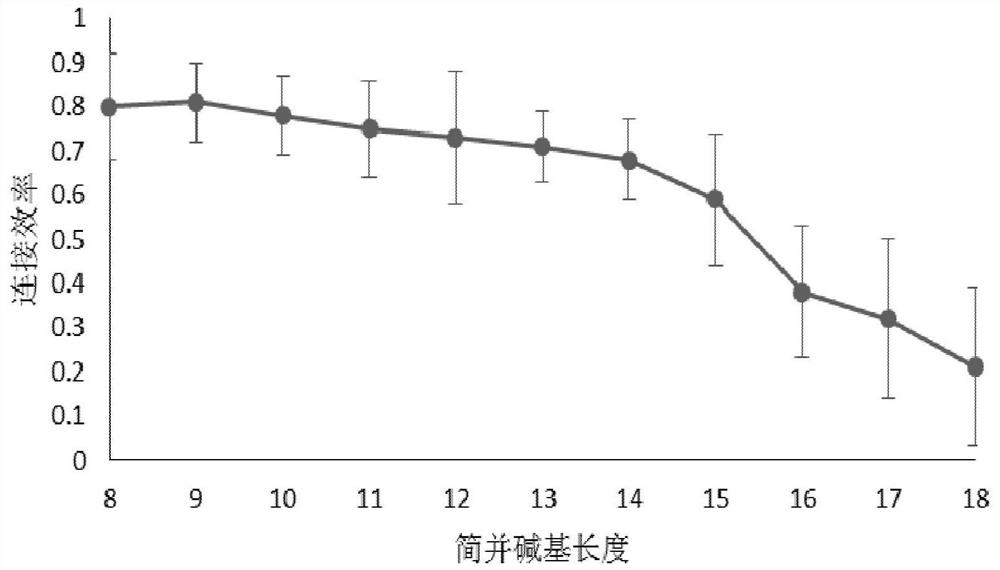
[0032] 5. Take a new PCR tube and add 1.1 μl adapter RA5 (where S2 is a degenerate base N with a length of 10 10 , S3 is selected from ACGA);
[0033] 6. Degenerate base N 10 The concentration was 10 μM, incubated at 70°C for 2 minutes, and placed on ice immediately after the reaction;
[0034] 6. Add 1.1 μl 10mM ATP, then add 1.1 μl T4RNA ligase and ...
Embodiment 2
[0055] 3' end plus connector
[0056] 1. Dilute the total RNA extracted from A549 cells with a total amount of 1 μg to a total volume of 5 μl with ultrapure water, and place it in a 200 μl thin-walled PCR tube;
[0057] 2. Add 1 μl adapter RA3, mix well, incubate at 70°C for 2 minutes, and immediately place on ice to cool;
[0058] 3. Add 2 μl HML (Ligation Buffer), 1 μl RNase Inhibitor, 1 μl T4RNA Ligase2, Deletion Mutant, mix well, and incubate at 28°C for 1 hour;
[0059] 4. Add 1 μl of STP (10mM Stop oligo solution) and mix well, incubate at 28°C for 15 minutes; add adapter at 5' end
[0060] 5. Take a new PCR tube and add 1.1 μl adapter RA5 (where S2 is a degenerate base N with a length of 15 15 , the concentration of N15 is 10 μM, S3 is selected from CCGA), incubated at 70°C for 2 minutes, and placed on ice immediately after the reaction;
[0061] 6. Add 1.1 μl 10mM ATP, then add 1.1 μl T4RNA ligase and mix well;
[0062] 7. Add 3 μl of the above Mix to the reaction ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com