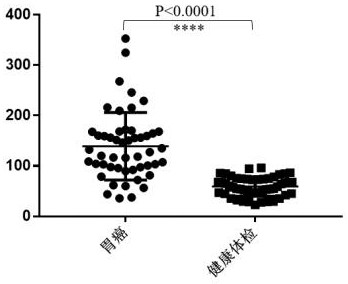
The application of the complex of cysteine protease inhibitor sn and cathepsin L1 as a diagnostic marker for gastric cancer
A cysteine protease and inhibitor technology, applied in the field of medical diagnostics, to achieve the effect of improving the detection rate and tissue specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
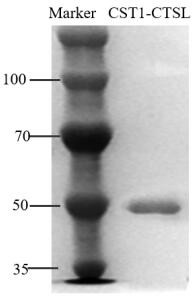
[0055] Expression and purification of embodiment 1CST1-CTSL recombinant protein
[0056] Protein expression: Synthesize the gene according to the amino acid sequence of SEQ ID NO: 1 in the sequence table and optimize the codons for mammalian expression. The gene was inserted into the pcDNA3.1 vector containing 6×His tag to obtain pcDNA3.1-CST1-CTSL. Then pcDNA3.1-CST1-CTSL was transformed into DH5α, and after positive clones were picked and cultured in large quantities, the recombinant plasmid pcDNA3.1-CST1-CTSL was extracted with a high-purity plasmid extraction kit. The recombinant plasmid was transferred into 293t cells, and the pcDNA3.1 empty vector was transfected at the same time as a negative control. 2 After culturing for 72 hours under the same conditions, the supernatant was collected and filtered with a 0.22 µm filter membrane.
[0057] SEQ ID NO:1MNPTLILAAFCLGIASATLTFDHSLEAQWTKWKAMHNRLYGMNEEGWRRAVWEKNMKMIELHNQEYREGKHSFTMAMNAFGDMTSEEFRQVMNGFQNRKPRKGKVFQEPLFYEAPRSV...
Embodiment 2
[0059] Example 2 Activity Verification and Antibody Pairing of CST1-CTSL Recombinant Protein
[0060] Activity analysis: Coat the chemiluminescence plate with 1ug / ml recombinant CST1-CTSL protein carbonate buffer (pH9.5) in a volume of 100ul at 4°C overnight, and serially dilute the capture antibody and enzyme-labeled antibody (concentration: 0~1ug / ml), add goat anti-mouse IgG-HRP (100ng / ml). The test found that the luminescence value of the capture antibody and the detection antibody at 100ng / ml was not less than 200,000, and the reaction curve between the protein and the antibody was R 2 >0.99, the reactivity of the protein meets the requirements.
[0061] Antibody pairing: Coat the chemiluminescence plate with 1ug / ml capture antibody, add 100ul of CST1-CTSL calibrator with different concentrations (5~1000pg / mL), incubate at 37°C for 60min, add 100uL of horseradish with a concentration of 100ng / ml after washing Peroxidase-labeled detection antibody, and incubated at 37°C ...
Embodiment 3
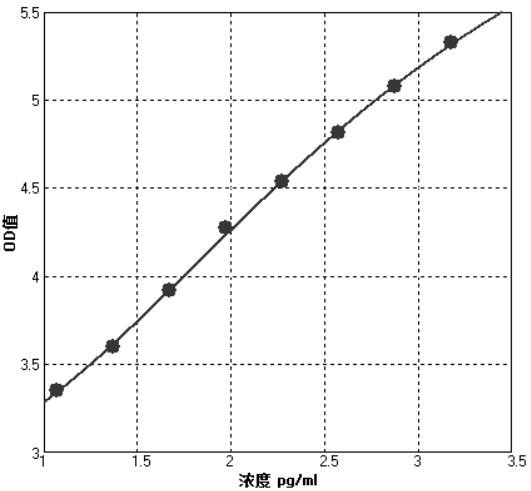
[0062] Embodiment 3 CST1-CTSL calibration curve drawing
[0063] Calibration curve drawing: First, coat the capture antibody on the chemiluminescence plate overnight at 4°C with a concentration of 1 μg / mL, and dilute the recombinant human CST1-CTSL calibrator protein with protein stabilizer to 0pg / mL, 10pg / mL, 50pg / mL , 100pg / mL, 200pg / mL, 500pg / mL, 1500pg / mL, 100μL per well, then add horseradish peroxidase-labeled detection antibody at a concentration of 5ng / ml, 100μL per well, and incubate at 37°C for 1 hour. Wash 3 times with PBST, add chemiluminescence substrate and measure the luminescence intensity of each well. The CST1-CTSL content of the tested samples was calculated from the calibration curve. The linear range of the calibration curve is 10~1500pg / mL, attached figure 2 It is the calibration curve of the CST1-CTSL detection kit, where the Y-axis represents the logarithmic value of the luminescence value, and the X-axis represents the logarithmic value of the concen...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
| affinity | aaaaa | aaaaa |
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com