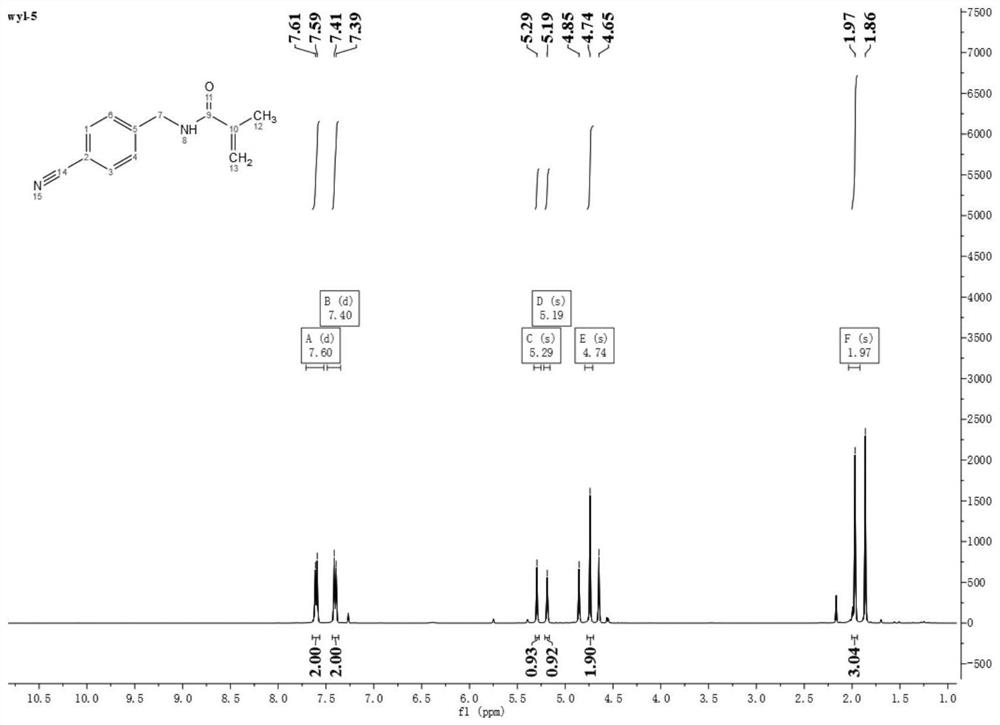
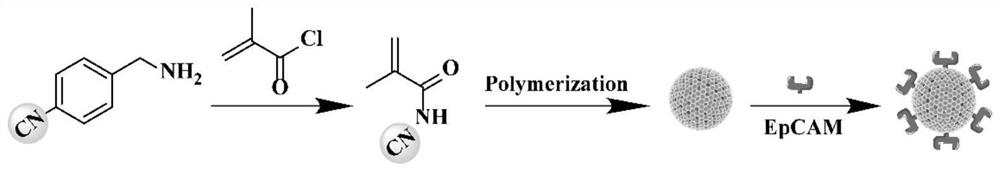
Micro-fluidic Raman chip and method for detecting exosome in blood based on micro-fluidic Raman chip
An exosome and microfluidic technology, applied in Raman scattering, chemical instruments and methods, biological testing, etc., can solve the problems of low stability and repeatability of immunoassay
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0057] A method for detecting exosomes in blood based on a microfluidic Raman chip. The method comprises the following steps:
[0058] 1. Preparation of exosome standard samples
[0059] Cell culture: LNCaP and normal prostate epithelial cells PrEC were cultured at 37°C in a medium containing RPMI 1640 (10% (v / v) fetal bovine serum and 1% (v / v) penicillin-streptomycin). When the growth reached 100%, the medium was removed, the cells were washed with PBS (2X), and the medium was collected after 72 hours of serum-starved culture (RPMI 1640, without fetal bovine serum).
[0060] Exosome extraction: Collect cell culture fluid, centrifuge at 4°C (3000g, 20min) to remove cells and cell debris, then filter with a 0.22μm filter to completely remove large cell debris, then centrifuge at 4°C, 120,000g for 60min , and the supernatant was removed by centrifugation twice. Finally, 100 μL of PBS was added to obtain the prepared exosome solution, which was stored at −20 °C for further anal...
Embodiment 2
[0070] Embodiment 2 Optimization of chip channel and flow rate
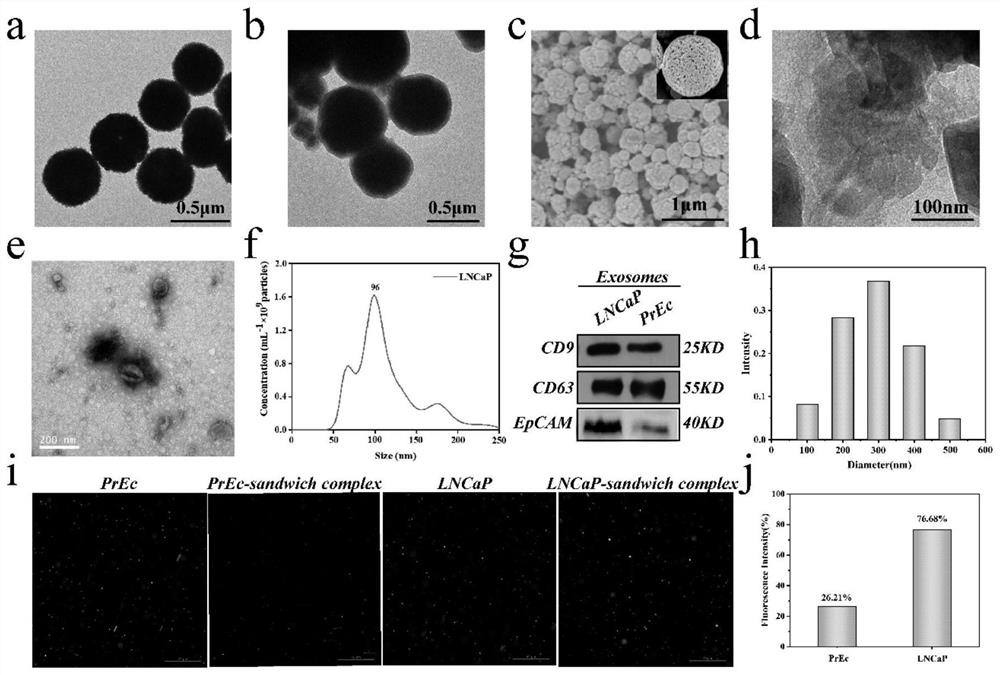
[0071] In order to optimize the micro-column spacing and flow rate in the mixing area, four spacings of 150 μm, 200 μm, 250 μm, and 300 μm were designed to study their impact on capture efficiency. Exosomes labeled with PKH26 staining flowed through the chip, and the fluorescence intensities of inflow and outflow were calculated. To calculate the capture efficiency of exosomes. Such as Figure 4 As shown in c, the capture efficiency of exosomes decreases with the increase of the spacing, to prevent the clogging of the microchannel caused by the aggregation of magnetic beads, we use 200 μm as the optimal spacing of the microcolumns. 4d is the flow rate optimization diagram, using 6 different flow rates for research, in order to quickly detect the target, choose 0.6μL min -1 For the optimal flow rate, the capture efficiency of exosomes was as high as 72.5% within 1 h.
[0072] designed as Figure 7 chip shown. ...
Embodiment 4
[0079] Example 4 Detection of Exosomes in Blood Samples
[0080] Serum samples were collected from 8 prostate cancer patients and 10 normal subjects. This experiment was approved by the Ethics Committee of Shanghai Ninth People's Hospital, and written informed consent was obtained at the beginning of the project. The details are shown in Table 1.
[0081] Table 1. Information on clinical serum samples.
[0082] Patient ID TNM sex Age, years PLT, 10 9 / L
[0083] Serum samples were filtered through a 0.22 μm filter, and then detected using the chip and method prepared in Example 1. Raman spectra were obtained by detecting a 20 μL sample accurately injected at a constant flow rate by a micro flow syringe pump. According to the previous detection of exosome samples, the content of exosomes in the actual blood sample was calculated, such as Figure 6 As shown in a, the exosome content values in the blood samples of prostate patients and normal people were...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com