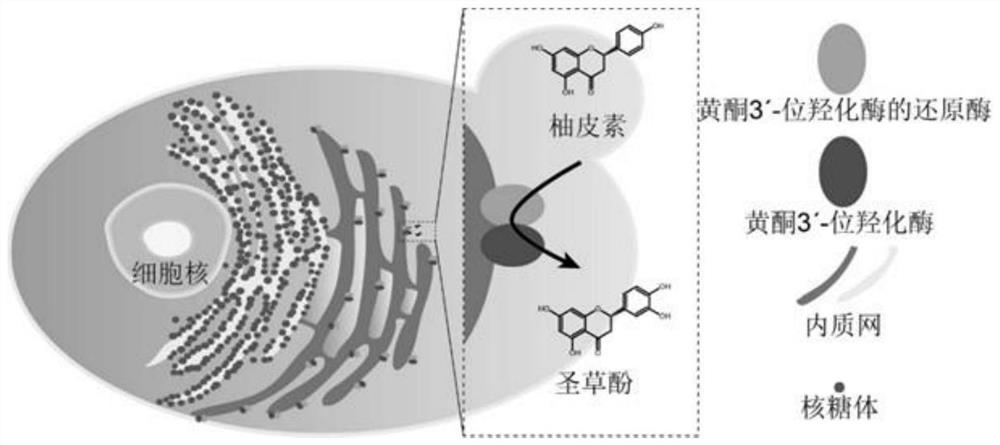
Application of flavone 3beta-hydroxylase from silybum marianum and coenzyme of flavone 3beta-hydroxylase
A technology of hydroxylase coenzyme and hydroxylase, which is applied in the field of genetic and metabolic engineering, can solve the problems of no activity, low expression activity, lack of efficient production, etc., and achieve the effect of increasing production
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0032] Example 1: Amplification and Characterization of SmF3'H and SmCPR
[0033] According to the kit instructions, total RNA was extracted from milk thistle stamens, and cDNA was obtained by reverse transcription. Using primers, respectively amplify Contig37917 (SmF3′H, using primers SmF3′H-F and SmF3′H-R) and Contig668 (SmCPR, using primers SmCPR-F and SmCP-R) from cDNA to obtain PCR products, and then the obtained PCR The products were cloned into pMD19T-simple to obtain pMD-T-SmCPR and pMD-T-SmF3'H respectively.
[0034]Because the original plasmid pY26-TEF-GPD has a PmlI restriction site, primers mut-F and mut-R were used to circularize and amplify the original plasmid pY26-TEF-GPF, and then caaggtttataa was obtained through self-ligation, and the plasmid pY26-TEF- GPD-mut introduces a new restriction site PmlI, and constructs pY26-TEF-GPD-mut for subsequent construction of plasmids.
[0035] The reported genes GhF3′H (GenBank ID: ABA64468.1) and CrCPR (GenBank ID: KM1...
Embodiment 2
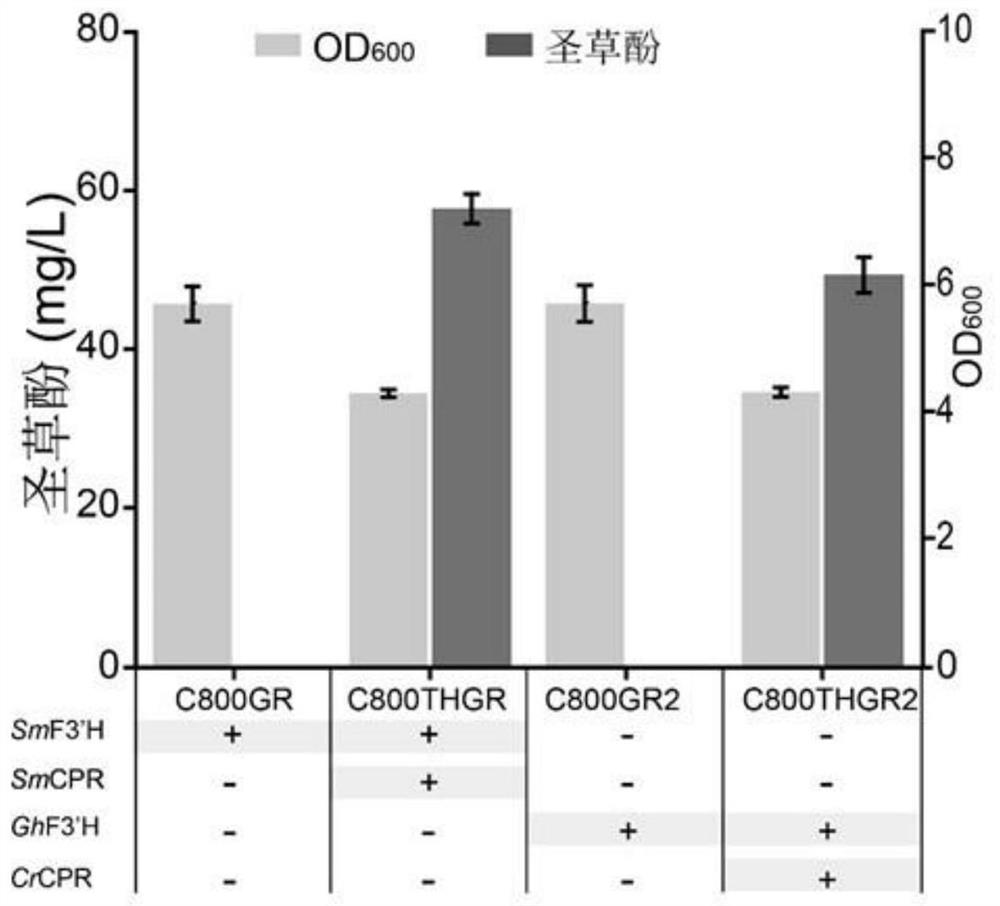
[0043] Example 2: Verification of the catalytic function of SmF3'H and SmCPR
[0044] Contig37917 (SmF3′H) and Contig668 (SmCPR) and strains C800GR, C800GR2, C800THGR and C800THGR2 were fermented in 250mL shake flasks. Conditions: Pick a single colony and inoculate in a 250mL shake flask containing 20mL YNB liquid medium, 30°C, 220rpm Cultivate for 16-18 hours to obtain seed medium. Transfer the seed medium at 2mL / 100mL to a 250mL shake flask containing 20mL of fresh YNB liquid medium containing 250mg L -1 Naringenin, 30°C, 220rpm cultured for 72 hours to detect the yield of eriodictyol.
[0045] The fermentation broth of strains Contig37917, Contig668, C800GR, C800GR2, C800THGR, and C800THGR2 was diluted with methanol to one-fold, shaken for 30s and mixed, centrifuged at 13500rpm for 5min and filtered to detect the yield of eriodictyol by HPLC. 250mL shake flask test: Mix 100μL fermentation medium with 900μL methanol and vortex for 30s, centrifuge at 13500rpm for 5min and f...
Embodiment 3
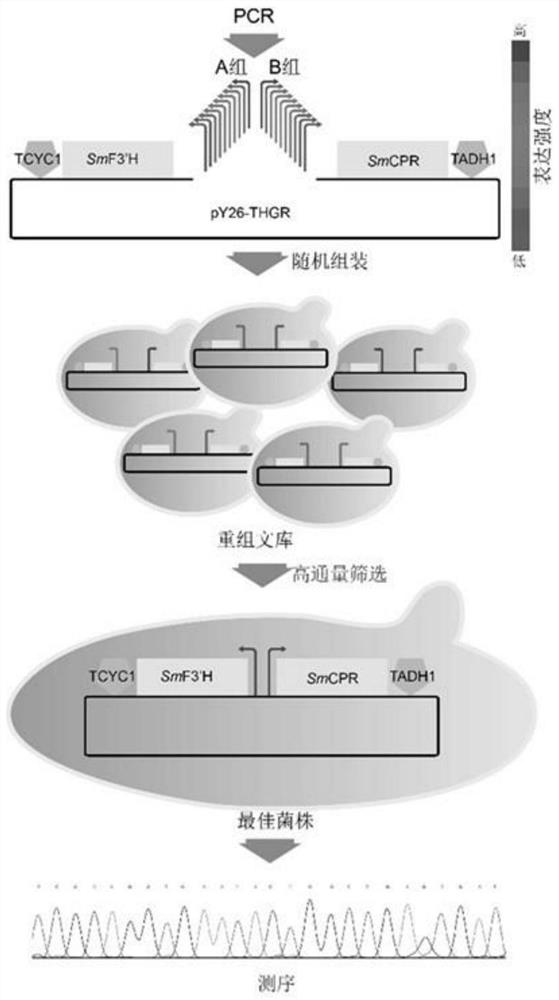
[0048] Example 3: Construction of promoter-optimized SmF3'H and SmCPR expression level libraries
[0049] From the reported promoter library (Gao, S., et al., Promoter-library-based pathway optimization for efficient(2S)-naringenin production from p-coumaric acid in Saccharomyces cerevisiae.J Agric Food Chem,2020.68(25):p .6884-6891.), 20 gradient promoters were selected for optimization of the expression levels of SmF3'H and SmCPR. The 20 promoters were divided into two groups, A and B. Promoter group A contains P INO1 ,P SED1 ,P TPI1 ,P MET6 ,P FAS2 ,P GAL1 ,P LEU2 ,P ZWF1 ,P ARO7 and P GLN1 (The sequence thereof corresponds to SEQ ID NO.5-SEQ ID NO.14 in turn), used for fusion and initial transcription of SmF3'H. Promoter group B contains P TDH1 ,P PGK1 ,P TDH3 ,P ERG20 ,P ADH6 ,P GAL10 ,P ADE2 ,P PMA1 ,P ADE6 and P FAD1 (Its sequence corresponds to SEQ ID NO.15-SEQ ID NO.24 in turn), used for fusion and initiation of transcription of SmCPR.
[0050] U...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com