Method for efficiently repairing gene mutation of ring-like sideroblastic anemia
A gene and mutation sequence technology, which is applied in the field of efficient repair of ring sideroblastic anemia gene mutations, can solve problems such as treatment options for unreported diseases, and achieve the effects of reconstruction, normal differentiation function, and promotion of differentiation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0175] Example 1: Efficient gene repair of ALAS-2 intron-1 point mutation in hiPSCs derived from XLSA patients
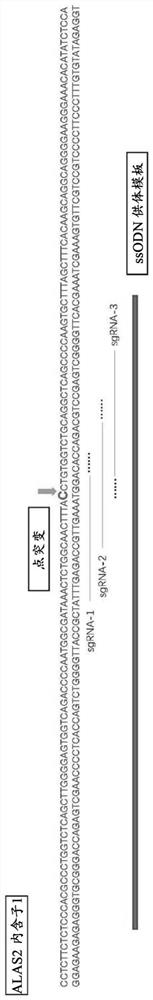
[0176] This example involves the use of the CRISPR / Cas9 system to gene-edit XLSA patient-derived human induced pluripotent stem cells (Human induced pluripotent stem cells, hiPSCs) to efficiently repair the ALAS-2 intron-1 point mutation at (X:55054635 [Chr X(GRCh37 / hg19):g.55054635A>G, NM 000032.4:c.-15–2187T>C), since this site is the junction of GATA-1 and ALAS-2 genes, this point is mutated Named Int-1-GATA.
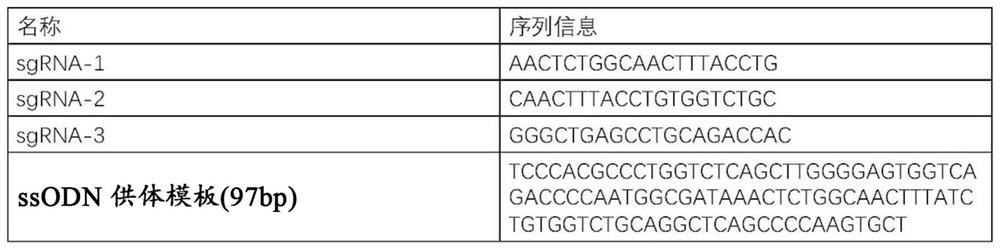
[0177] In order to repair disease mutations, first use the "CRISPR RGEN TOOLS" software to design sgRNA targeting the genome near the Int-1-GATA mutation site and synthesize three chemically modified sgRNAs, wherein the sgRNA contains a sequence complementary to the target sequence The coding information is as follows: sgRNA-1: aactctggcaactttacctg (SEQ ID NO: 1), sgRNA-2: caactttacctgtggtctgc (SEQ ID NO: 2), sgRNA-3: gggctgagcctgcagaccac (SEQ ID NO: 3)...
Embodiment 2
[0189] Example 2 Efficient Gene Repair of ALAS-2 Intron-1 Point Mutation in CD34+ HSPCs from Bone Marrow of XLSA Patients
[0190] In Example 1, we achieved efficient repair of the Int-1-GATA point mutation of XLSA-derived hiPSCs, referring to the amount of Cas9 mRNA, sgRNA-1 and ssODN added in Example 1, in this experiment we tried gene repair CD34+ HSPC derived from bone marrow of XLSA patients.
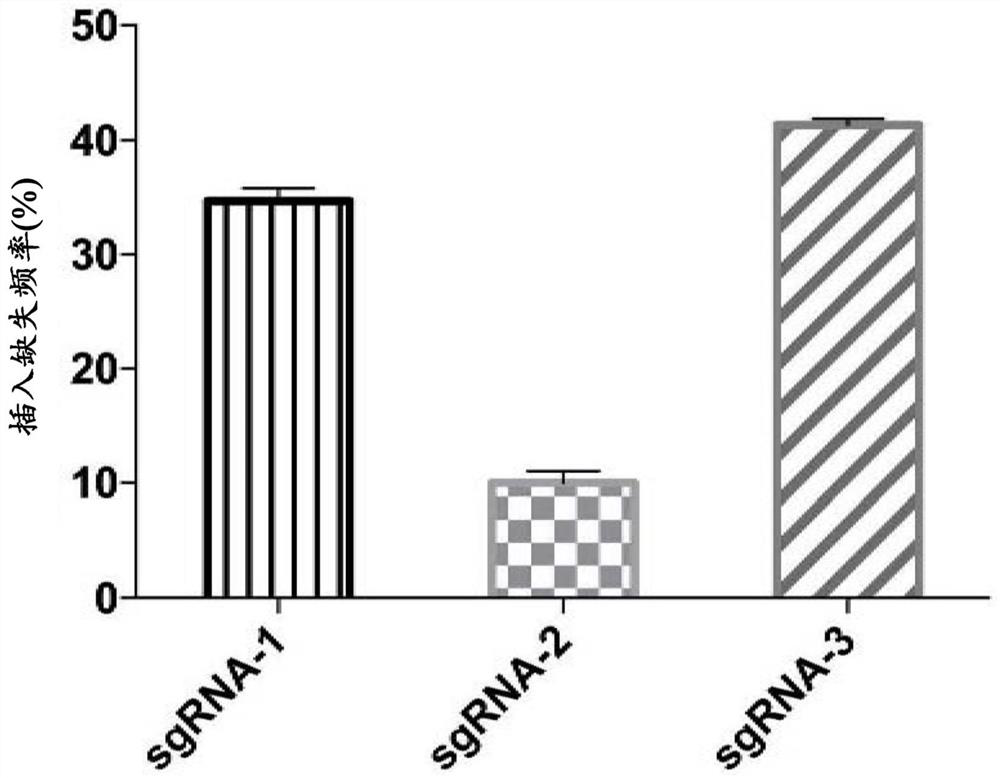
[0191] Select the electroporation conditions of 300v 1ms, electrotransfer the amount of different Cas9 mRNA, sgRNA-1 and ssODN into the CD34+HSPC derived from the bone marrow of XLSA patients, and the Cas9 mRNA:sgRNA-1:ssODN added in every 1.0*10^6 cells is 6 μg: 4 μg: 6 μg and 6 μg: 4 μg: 10 μg, after 4 days, the genome of the HSPC was extracted, and the gene repair efficiency (HDR) and Indels efficiency (NHEJ) were analyzed by next-generation sequencing bioinformatics methods, such as Image 6 shown. The results showed that under the conditions of different addition amounts o...
Embodiment 3
[0193] Example 3 CD34+HSPC derived from XLSA patient's bone marrow after erythroid differentiation gene repair to evaluate cell phenotype Change
[0194] 3.1 Red blood cell differentiation
[0195] Referring to the amount of Cas9 mRNA, sgRNA-1 and ssODN found in Example 2 (6 μg: 4 μg: 12 μg), select the electroporation condition of 300v 1ms, and respectively electroporate Cas9 mRNA, sgRNA-1 and ssODN into the HSPC derived from the bone marrow of XLSA patients , erythroid differentiation experiments were performed using the "two-step" differentiation protocol described below. In addition, we erythroid differentiated CD34+ HSPCs from peripheral blood mobilized from healthy donors as a positive control.
[0196] The two-step method is to use HSPC erythroid expansion and differentiation medium for differentiation, and then use HSPC erythroid differentiation enucleation medium for differentiation.
[0197] Hematopoietic Stem Cell Erythroid Expansion and Differentiation Medi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com