Synthesis method of target gene segment
A gene fragment and synthesis method technology, applied in the field of gene fragment synthesis, can solve the problems that primers cannot anneal and extend each other, reduce synthesis flux, and increase synthesis cost, so as to save cycle and manpower input, synthesis efficiency and flux High, ensure the effect of synthesis cycle
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
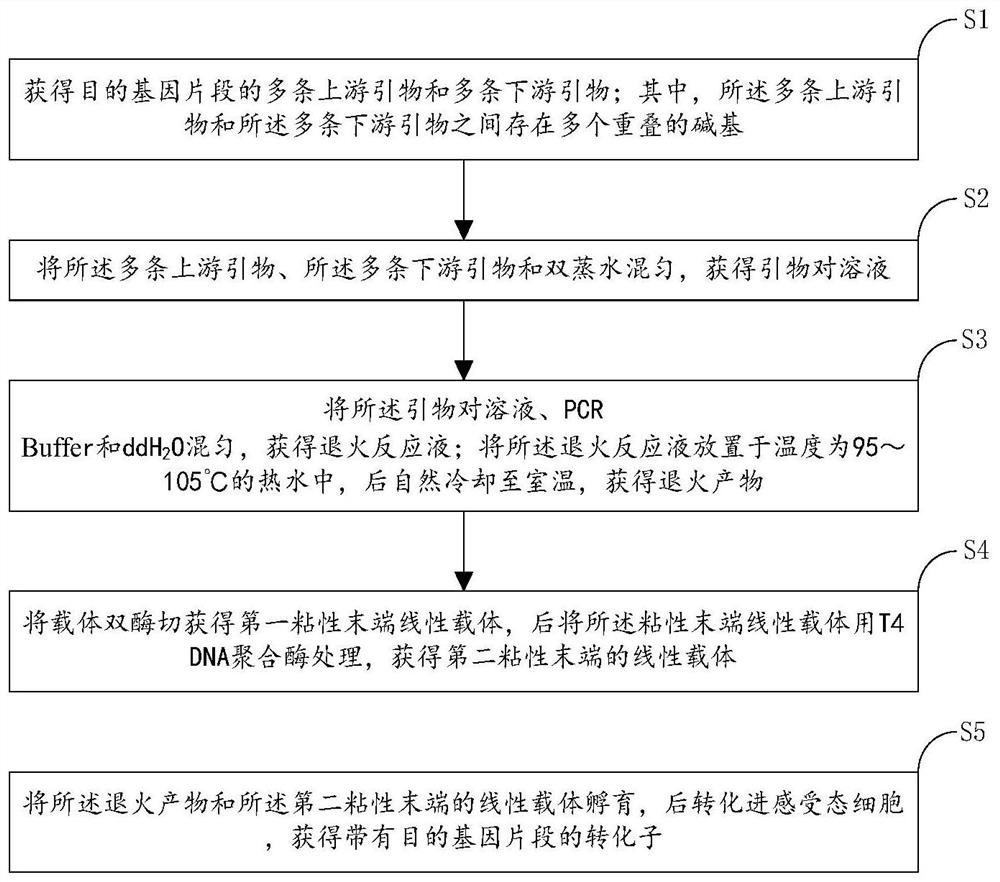
[0034] According to a typical embodiment of the present invention, a method for synthesizing a target gene fragment is provided, such as figure 1 As shown, the method includes:
[0035] S1. Obtain multiple upstream primers and multiple downstream primers for the target gene fragment; wherein, there are multiple overlapping bases between the multiple upstream primers and the multiple downstream primers;
[0036] S2. Mix the multiple upstream primers, the multiple downstream primers and double distilled water to obtain a primer pair solution;
[0037] S3, the primer pair solution, PCR Buffer and ddH 2 O mixing to obtain an annealing reaction solution; placing the annealing reaction solution in hot water at a temperature of 95-105° C., and then naturally cooling to room temperature to obtain an annealing product;
[0038] S4. Digesting the vector with double enzymes to obtain a linear vector with a first sticky end, and then treating the linear vector with a sticky end with T4 ...
Embodiment 1
[0057] 1. Experimental materials
[0058] 1. Main experimental instruments
[0059] Table 1
[0060]
[0061] 2. Main experimental reagents
[0062] Table 2
[0063]
[0064] 2. Experimental steps
[0065] 1. Design annealing primers:
[0066] table 3
[0067]
[0068]
[0069] The 190186B-1-M, 190186B-3-M, 190186B-5-M, 190186B-7-M, 190186B-9-M and 190186B-11-M are upstream primers; the 190186B-2-M, 190186B -4-M, 190186B-6-M, 190186B-8-M, 190186B-10-M and 190186B-12-M are upstream primers; among them, the overlap between upstream primers and downstream primers has been listed in Table 3 Marked in black font;
[0070] Mix the upper and lower primers, add 100 ul of double distilled water to dissolve, and obtain a primer pair solution; the upper and lower primers are selected from the two in Table 3, and the final primer concentration is 50 pmol / μl.
[0071] 2. Prepare the following reaction system in the PCR tube, and the final reaction system is 20ul (as sho...
Embodiment 2
[0082] 1. Design annealing primers:
[0083] Table 7
[0084]
[0085] The 190779A-1-M, 190779A-3-M, 190779A-5-M, 190779A-7-M and 190779A-9-M are upstream primers; the 190779A-2-M, 190779A-4-M, 190779A -6-M, 190779A-8-M and 190779A-10-M are downstream primers. Among them, the overlap between upstream primers and downstream primers has been marked in black font in Table 7;
[0086] Mix the upper and lower primers, add 100 ul of double distilled water to dissolve, and obtain a primer pair solution; the upper and lower primers are selected from the two in Table 7, and the final concentration of the primers is 50 pmol / μl.
[0087] 2. Prepare the following reaction system in the PCR tube, and the final reaction system is 20ul (as shown in Table 8). Put the PCR tube into freshly boiled water and let it cool down to room temperature naturally.
[0088] Table 8
[0089]
[0090] 3. Mix 1-2 μg of the carrier (the nucleotide sequence of the carrier is shown in SEQ ID NO: 13), 5 ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com