Rapid detection kit for snake source components and application of rapid detection kit
A snake-derived, rapid technology, applied in the field of biological species identification, can solve the problems of difficult identification and achieve the effect of low copy number, easy observation and high sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0049] Example 1 Snake source universal internal standard gene JUN screening
[0050] The JUN (Gen bank No. EF144048.1 ) gene was screened out by searching the gene information about snakes in Gen Bank, and using BLAST software for homology and specificity analysis.
[0051] The internal standard gene screening process is as follows:
[0052] Step 1. Query the genome information of the target species in the NCBI database, analyze its integrity, select and download the species genome with complete sequencing information, and store it in the ".FASTA" format. Species that need to be screened and distinguished from the target species are limited, that is, "compared species", which are generally common species that are closely related to the target species. The selection of comparison species will directly affect the speed and accuracy of screening comparison, that is, the number of comparison species is large and the comparison speed is slow, so it is necessary to choose flexib...
Embodiment 2
[0064] Example 2 Establishment of LAMP detection method for snake source components
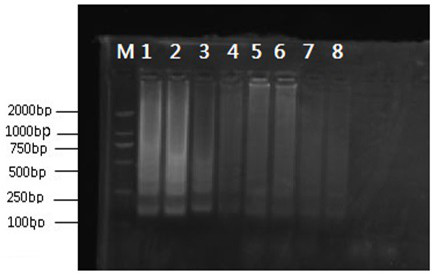
[0065] against JUN Gene (Genbank No. EF144048.1), using the Japanese Rongyan Co., Ltd. ring-mediated primer online design software, a total of 4 pairs of primers were designed, named JUN-1, JUN-2, JUN-3, JUN-4 (Table 1). Utilize these 4 pairs of primers to carry out LAMP amplification to snake source genomic DNA, and use 2% agarose gel to carry out electrophoresis analysis to the amplified product ( figure 2 ), the results showed that the band of primer JUN-1 was the clearest and brightest, and JUN-1 was selected for subsequent experiments.
[0066] Table 1 LAMP primer sequence
[0067]
[0068] Use LAMP to quickly detect snake samples, the reaction system is 25 μL, including 1×Bst Thermal buffer, 0.6mM dNTP, 3.6 mM MgSO 4 , 0.6 M betaine, 1.6 μM primers JUN - 1-FIP, 1.6 μM primer JUN - 1-BIP, 0.2 μM primer JUN -1-F3, 0.2 μM primer JUN - 1-B3, 8 U Bst DNA polymerase large fra...
Embodiment 3
[0070] Example 3 Universal identification of the standard gene LAMP in snakes
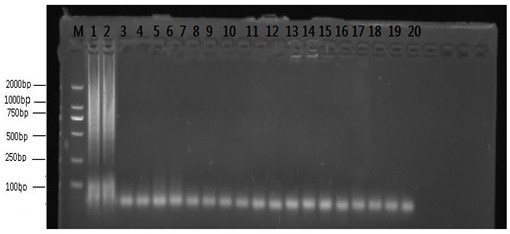
[0071]This application uses the designed primers JUN-F / R to carry out LAMP amplification on three kinds of snakes and use 2% agarose gel to perform electrophoresis analysis on the amplified products. The results showed that the snake meat of the three species had bright amplification bands ( Figure 4 ). This indicates that the gene and corresponding primers can be used to detect snake-derived products universally.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com