Recombinant yeast chassis cell transformation for efficiently converting chenodeoxycholic acid and construction and application of recombinant strain
A technology of chenodeoxycholic acid and yeast strains, applied in the field of biosynthesis, can solve problems such as ambiguity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
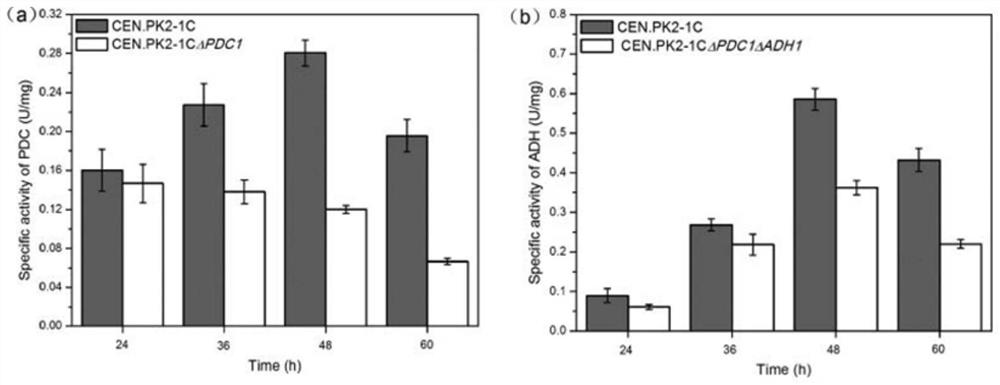
[0029] Example 1: Construction of mutant yeast strain S.cerevisiae CEN.PK2-1CΔpdc1Δadh1
[0030] Knockout of the PDC1 gene using Cre-LoxP technology:
[0031] Using the plasmid pUG27 with the HIS tag as a template, PDC1-F and PDC1-R as primers (the primer sequences are shown in SEQ.NO.05-SEQ.NO.06), PCR amplification obtained 1544bp for knocking out PDC1 The HIS knockout box of the gene includes the HIS gene and the 45 bp nucleotide homologous sequence between the upstream and downstream of the PDC1 gene. The wild-type Saccharomyces cerevisiae strain CEN.PK2-1C was transformed by LiAc transformation method, coated with SD-His plate, and cultured in a 30°C incubator for 3 days. The obtained transformant was streaked and purified on the SD-His plate for 2-3 days, then inserted into the SD-His liquid medium and cultured overnight to saturation, and the genome was extracted for PCR verification. The primers A (PDC1), BM (PDC1), The corresponding sequences of CM(PDC1) and D(PDC1)...
Embodiment 2
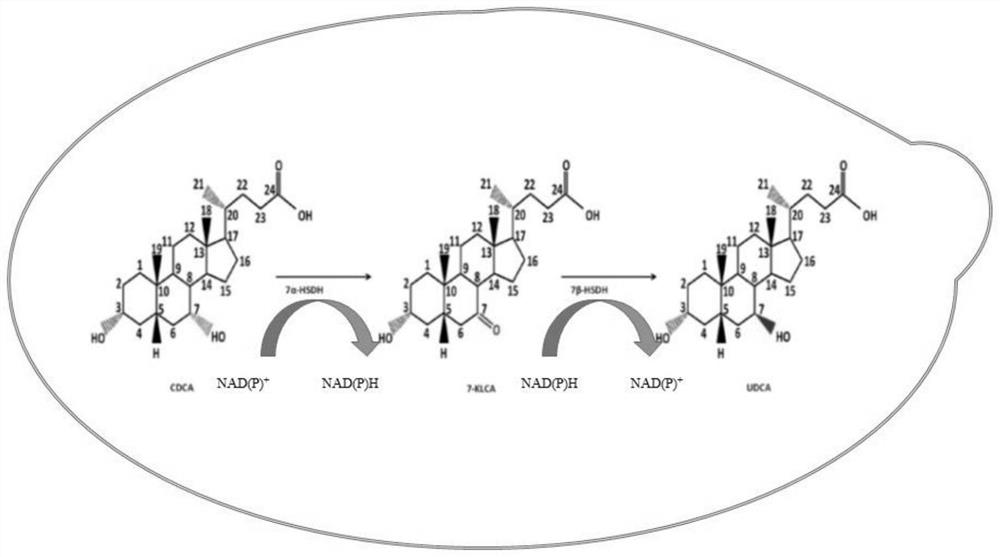
[0037] Example 2 Construction of Transformed Chenodeoxycholic Acid Engineering Yeast
[0038] Using pY15TEF1 and pYX212 as expression plasmids, construct pY15TEF1-7α-HSDH and pYX212-7β-HSDH overexpression vectors. The specific method is as follows: the gene products 7α-HSDH and 7β-HSDH with restriction enzyme sites were synthesized by Shanghai Sangon Bioengineering Technology Service Co., Ltd., and then the above gene products 7α-HSDH and 7β-HSDH were combined with the plasmid pY15TEF1 , pYX212 were simultaneously digested with XbaI and BamHI, EcoRI and BamHI (primer sequences are shown in SEQ.NO.17-SEQ.NO.20), ligated with T4 DNA ligase, and 5 μL of the ligated product was transformed into 50 μL Escherichia coli competent Cells were spread on LA plates, and the correctness of the obtained transformants was verified by colony PCR and enzyme digestion, double enzyme digestion with XbaI and BamHI, EcoRI and BamHI was used to verify the correct transformants, and the recombinant ...
Embodiment 3
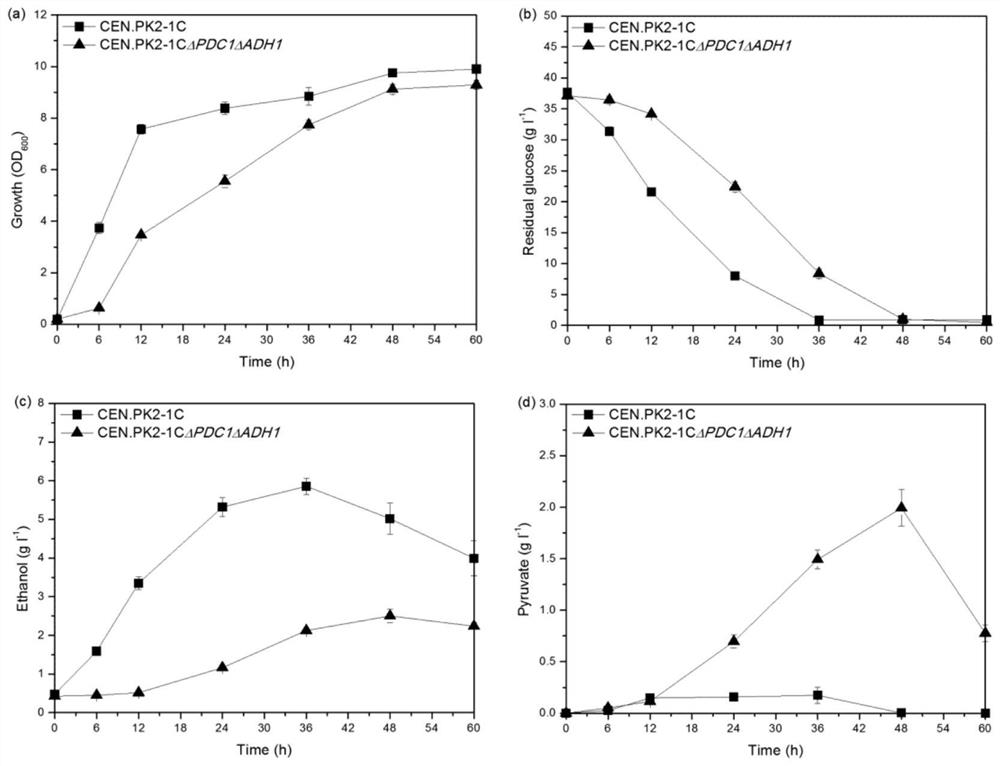
[0042] Embodiment 3 Fermentation of transformed chenodeoxycholic acid engineered yeast
[0043] Saccharomyces cerevisiae culture conditions: Bacteria were taken from the strain preservation tube at -80°C, activated on YPD (transformants containing nutrient marker plasmids in the corresponding SD-deficient medium) plates, and cultured at 30°C for 3 days; pick the activated single colony and inoculate Into a 3mL test tube containing YPD (transformants containing nutrient marker plasmids in the corresponding SD-deficient medium) medium, cultivate overnight at 30°C and 220rpm until saturated.
[0044] Fermentation culture conditions: Pick an activated single colony from the plate and inoculate it into the seed medium, and culture it in a shaker flask at 30°C and 220rpm for 24h until saturated. Transfer to the fermentation medium shake flask with initial OD=0.2, culture at 30°C and 220rpm. The cells were cultured to the stationary phase (48h), collected by centrifugation, resuspen...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com