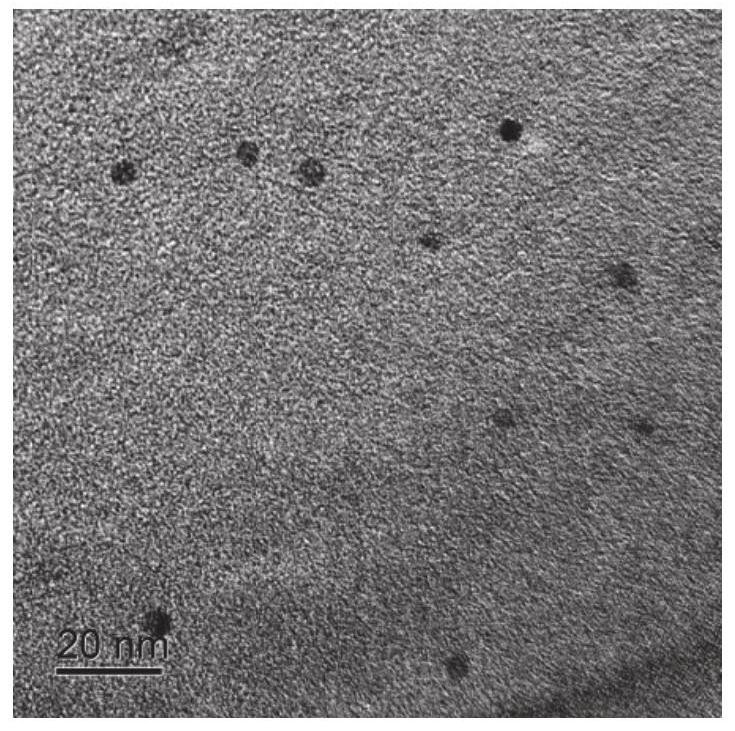
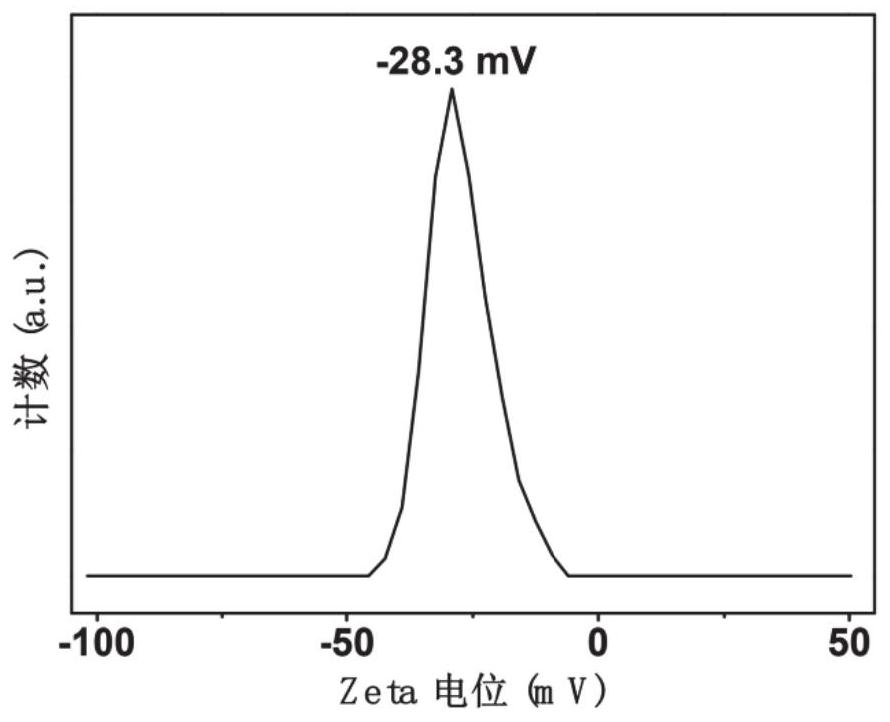
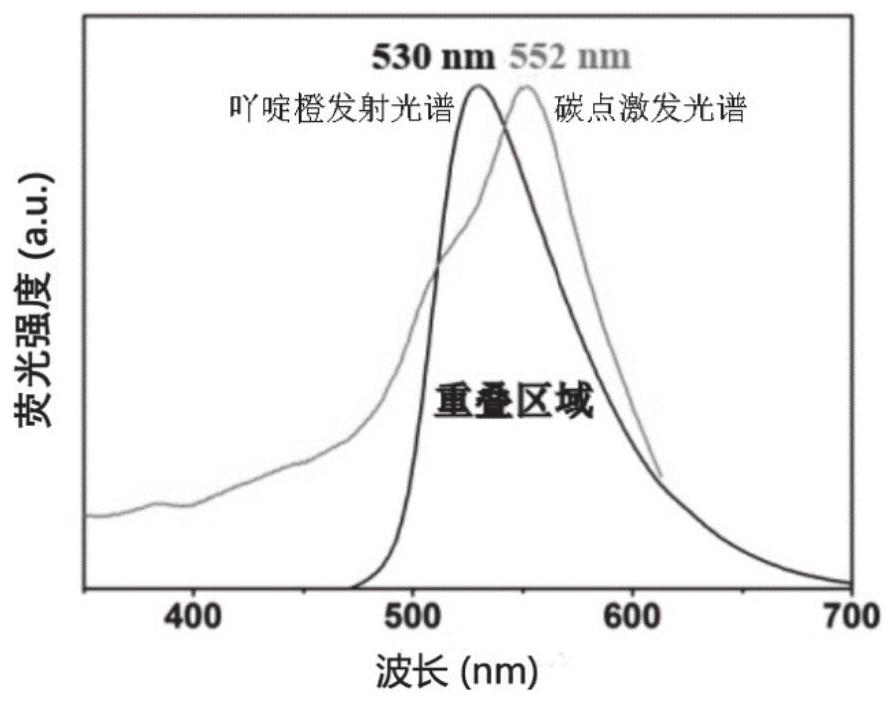
A method based on ratiometric fluorescence of acridine orange and carbon dots for miRNA detection in vitro
A ratiometric fluorescence, in vitro detection technology, applied in the field of miRNA in vitro detection, can solve the problems of complex components, background fluorescence interference system, quenching, etc., and achieve the effect of good repeatability and broad application prospects.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0043] Preparation method of carbon dots: Dissolve 1.8g citric acid in 30mL formamide and transfer to a polytetrafluoroethylene-lined stainless steel reaction kettle, react at 180°C for 6 hours, and pour into the reaction solution after the reaction kettle is naturally cooled to room temperature Add 120mL of acetone and put it in the refrigerator, place it at -20°C for 24 hours to produce carbon dot precipitation, centrifuge at 10,000 rpm for 20 minutes to separate the carbon dot precipitation, add the carbon dot precipitation to 30mL methanol / acetone with 10% methanol volume In the mixed solution, wash by centrifugation at 10,000 rpm for 20 minutes, repeat the centrifugation and washing 3 times, redissolve the carbon dots in 10 mL of methanol, filter with a filter membrane with a pore size of 0.22 μm to remove large particles, and pour the carbon dots into the methanol solution. Add 30 mL of acetone, centrifuge and wash at 10,000 rpm for 20 minutes, and dry at 50°C for 12 hour...
Embodiment 2
[0048] Add 3 μL of miRNA solution to 292 μL of DNA probe solution with a concentration of 10 nM, so that the final concentration of miRNA is 8.5 nM, incubate at 37 ° C for 90 minutes, add 3 μL of acridine orange solution with a concentration of 1.76 μM and adsorb for 10 minutes, Then add 2 μL of carbon dot solution with a concentration of 0.1 mg / mL and let it stand for 10 minutes, and test the fluorescence of the solution with a fluorescence spectrophotometer, according to Figure 5 The standard curve equation in calculates the concentration of miRNA as 8.43nM, which is close to the actual concentration of miRNA. The test results show that the detection method has high accuracy.
Embodiment 3
[0050] Add 3 μL of three control miRNAs (sequence from 5' to 3', No.1(hsa-miR-223-3p): UGUCAGUUUGUCAAAUACCCCA, No.2(miR- 92a-3p-TMT): UUUUCCACUUGUCCCGGCCUGU, No.3 (miR-92a-3p-SMT): UAUUCCACUUGUCCCGGCCUGU) solution to make the final concentration of control miRNA 7nM, incubate at 37°C for 90 minutes and add 3 μL to it at a concentration of 1.76 μM acridine orange solution and adsorb for 10 minutes, then add 2 μL carbon dot solution with a concentration of 0.1 mg / mL and let it stand for 10 minutes, and test the fluorescence of the solution with a fluorescence spectrophotometer, according to Figure 5 The standard curve equation in calculates the concentrations of the three control miRNAs as -0.12, -0.19 and 0.01nM, which are close to 0. The detection results showed that the detection method had good specificity.
PUM
| Property | Measurement | Unit |
|---|---|---|
| concentration | aaaaa | aaaaa |
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com