Colorectal cancer targeted therapy drug
A colorectal cancer and drug technology, applied in the field of tumor cell biology, can solve the complex problems of colorectal cancer, achieve excellent diagnostic value and inhibit proliferation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
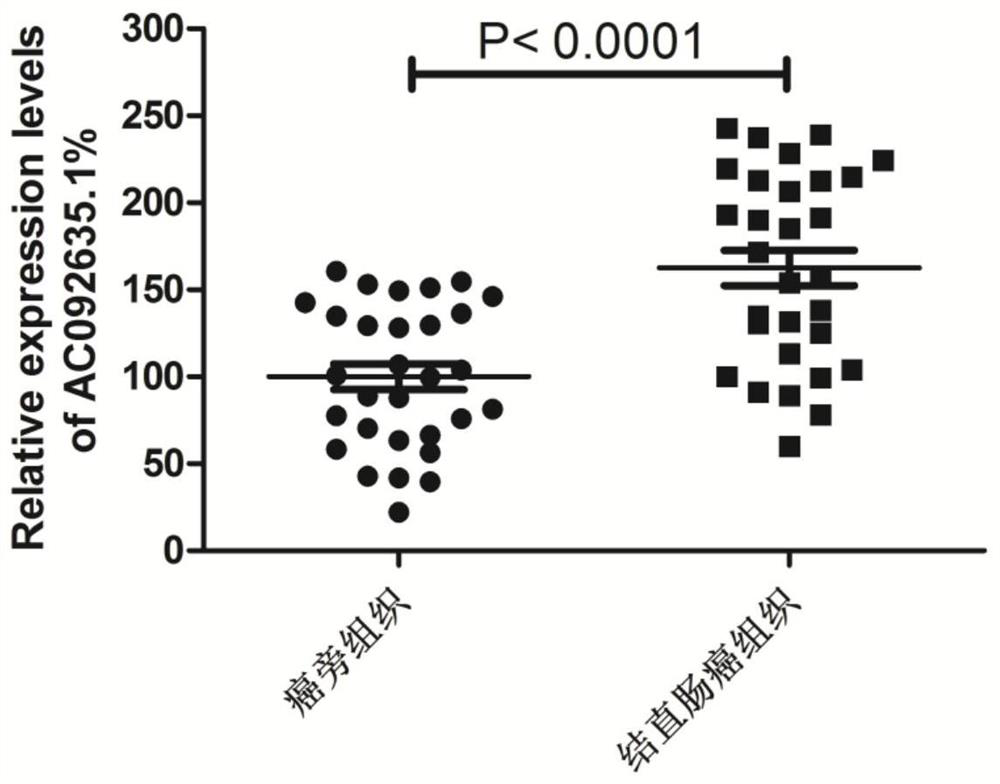
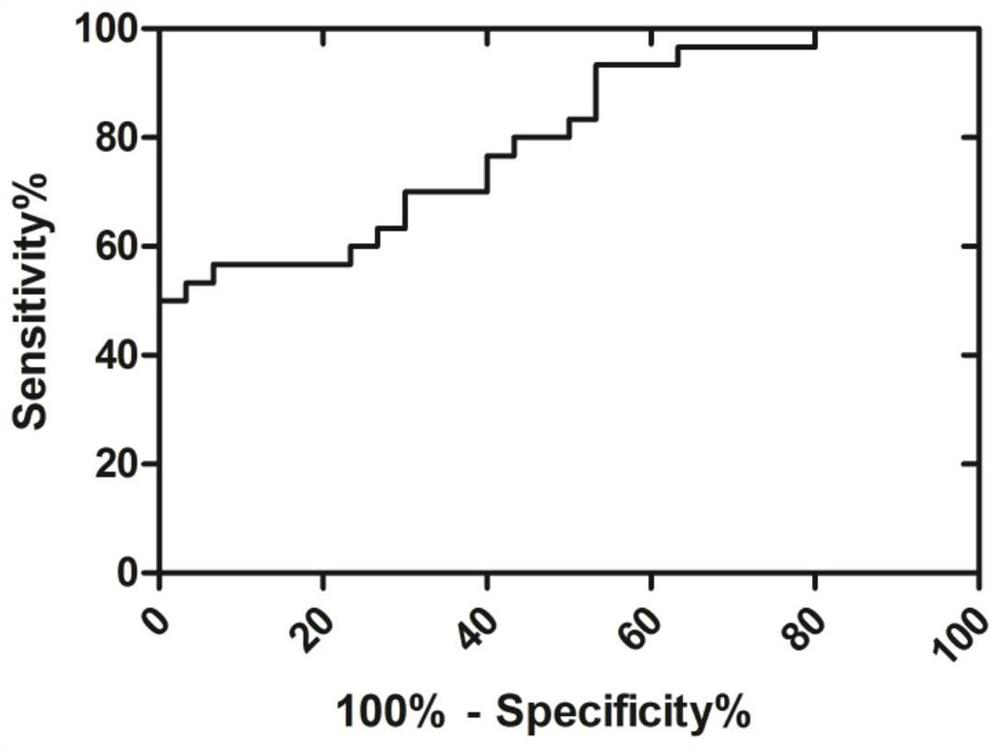
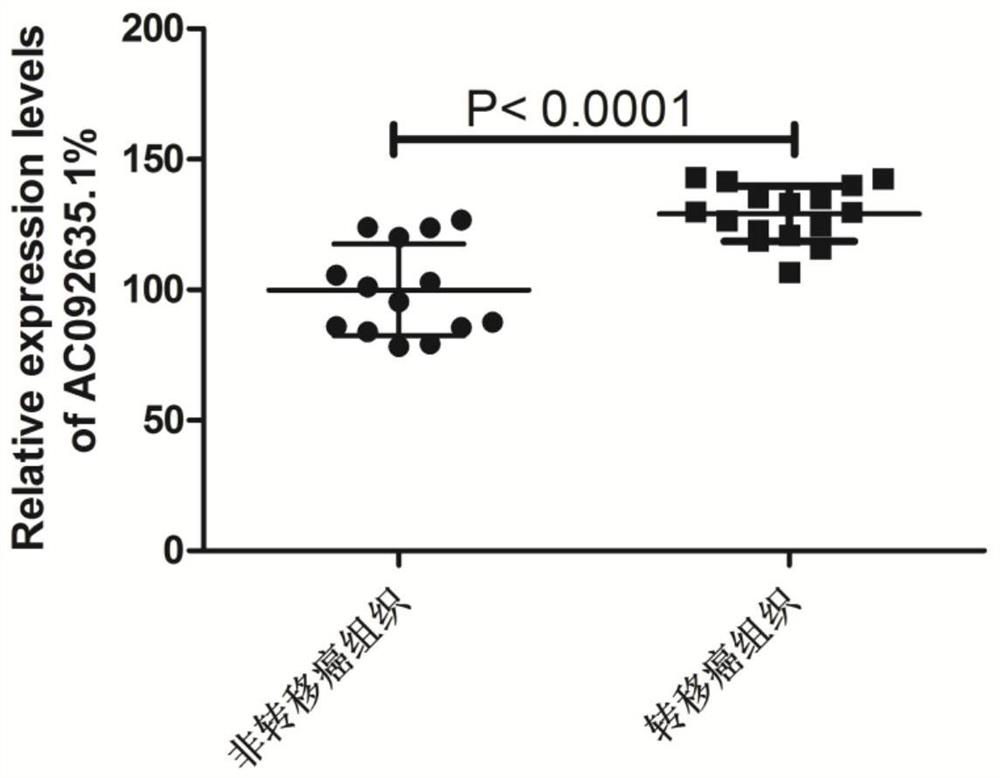
[0035] Detection of AC092635.1 Gene Expression Differences in Colorectal Tumor Tissues and Paracancerous Tissues
[0036] 1. Extraction of RNA from Tissues
[0037] (1) Put 50mg of colorectal cancer tissue and paracancerous tissue into a pre-cooled mortar, and quickly grind it into powder;
[0038] (2) Add 1ml Trizol to the mortar, mix thoroughly, transfer to an enzyme-free centrifuge tube, and let stand at room temperature for 5 minutes;
[0039] (3) Set the speed of the centrifuge to 12000rpm, put it into a centrifuge tube and centrifuge for 5min;
[0040] (4) Add 200 μl chloroform to the centrifuge tube, mix it upside down, and let it stand at room temperature for 10 minutes;
[0041] (5) Set the speed of the centrifuge to 12000rpm, put it into the centrifuge tube and centrifuge for 10min;
[0042] (6) The liquid is divided into three layers, transfer the supernatant of the upper layer to a new enzyme-free centrifuge tube, add an equal volume of pre-cooled isopropanol to...
Embodiment 2
[0072] Detection of the expression difference of AC092635.1 in colorectal cancer cells by fluorescent quantitative PCR
[0073] (1) Inoculate human normal colonic epithelial cells NCM460, HT29, SW620, SW480 and LOVO in the logarithmic growth phase in cell culture plates;
[0074] (2) After the cell density reaches 90%, add Trizol to extract RNA, RNA extraction, reverse transcription reaction and fluorescence quantitative PCR reaction steps are the same as in Example 1.
[0075] Differences in the expression of AC092635.1 in different colorectal cancer cells are as follows: Figure 5 shown. It can be seen that the expression level of AC092635.1 in colorectal cancer cells is significantly higher than that in normal human colonic epithelial cells NCM460, which is consistent with the expression level in tissues.
Embodiment 3
[0077] Design the interference RNA of AC092635.1 and verify the interference effect
[0078] (1) Design siRNA according to the sequence ENST00000434509.1 of the AC092635.1 gene, and the sequence of si AC092635.1 is as follows:
[0079] Justice strand: AAUUUUCCCCCUUUCUGUGGG, SEQ ID NO.6
[0080] Antisense strand: CACAGAAAGGGGGAAAAUUAG, SEQ ID NO.7;
[0081] (2) Inoculate SW480 cells in a 6-well plate. When the cells grow to 80%, transfect siNC and siAC092635.1 according to the instructions of LipoFiterTM 3.0. After 48 hours of transfection, extract RNA for fluorescent quantitative PCR detection. For specific steps, see Implementation example 1.
[0082] The knockdown effect of siAC092635.1 is as follows Figure 6 As shown, among them, the relative expression level of AC092635.1 in the siAC092635.1 group cells was (27.30±6.02)%. It can be seen that siAC092635.1 can effectively inhibit the expression of AC092635.1.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com