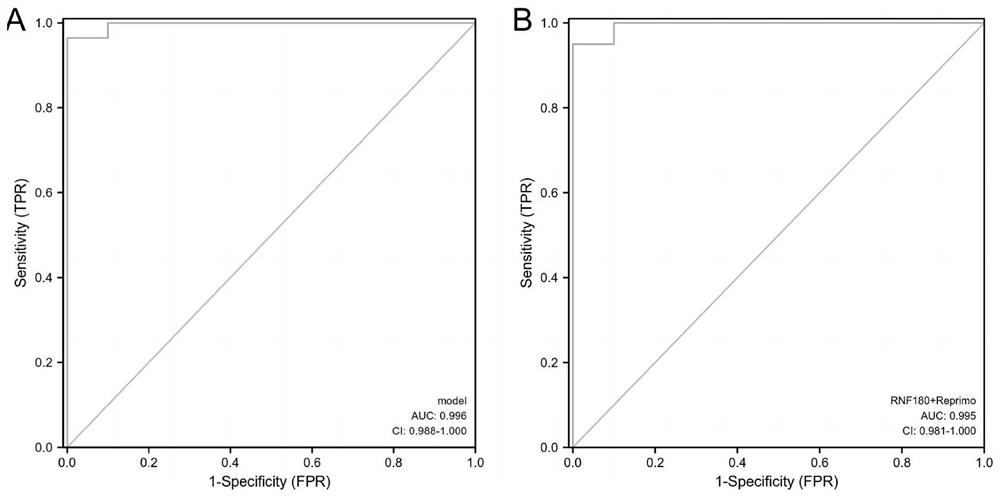
Composition for detecting free DNA methylation of plasma based on digital PCR and application of composition
A technology of methylation and composition, which is applied in the field of detection of plasma free DNA methylation composition, can solve the problems of inaccurate clinical detection of gastric cancer, difficulty in realizing high-precision and absolute quantitative detection of plasma DNA methylation, etc. , to achieve the effect of solving tissue sample dependence, broad application prospects and industrialization prospects, and saving time
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0035] (1) Extraction of plasma cfDNA
[0036] 1) Add 30 μL of magnetic bead suspension, 55 μL of proteinase K, and 150 μL of Bead BindingBuffer to 1 mL of plasma sample in sequence, and vortex to mix;
[0037] 2) Centrifuge at 200×g for 30s to remove the liquid on the lid, place the Ep tube on a 2mL magnetic stand, incubate for 1min until the solution is clear, and remove the upper layer solution with a pipette;
[0038] 3) Remove the Ep tube, add 200 μL Bead Elution Buffer to it, vortex to mix, transfer the mixture to the Bead Elution tube, put it in the metal bath, set the program at 20°C, 300 rpm, and incubate for 5 minutes;
[0039] 4) Remove it and place it on a magnetic stand for 1 min until the solution is clear. Transfer the supernatant to a new elution tube, discard the magnetic beads, add 300 μL ACB buffer to the supernatant, vortex to mix, and briefly centrifuge;
[0040] 5) Add the mixture from the previous step to the QIAamp UCP Elution Column, centrifuge at 60...
Embodiment 2
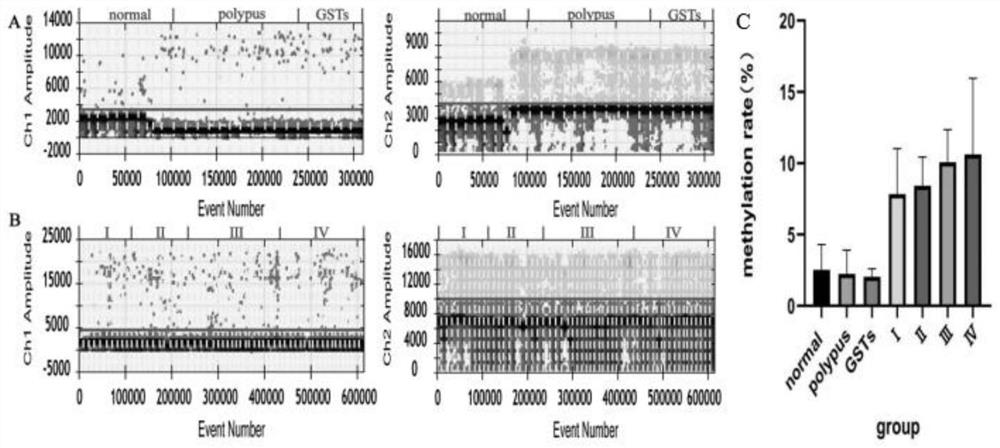
[0056] Design of primers for digital PCR amplification: The methylated sequence and non-methylated sequence of the gene Reprimo bisulfite conversion were used as the target gene to design primers. The primer sequences are listed in Table 1.
[0057] Table 1 Primer sequence list
[0058]
[0059]
[0060] Among them, the fluorophore labeled at the 5' end of M-Probe is FAM, the quencher group labeled at the 3' end is BHQ1; the fluorophore labeled at the 5' end of U-Probe is VIC, and the quencher group labeled at the 3' end is The killing group is BHQ1.
[0061] 1) Assuming that the sample to be tested is N, the configuration of the reaction system is shown in Table 2:
[0062] Table 2 ddPCR reaction system
[0063]
[0064] 2) After fully mixing the reaction system, dispense it into 17 μL of each reaction tube;
[0065] 3) Take 3 μL of bisulfite-converted DNA and add them to corresponding reaction tubes, with a final volume of 20 μL;
[0066] 4) Put a new droplet g...
Embodiment 3
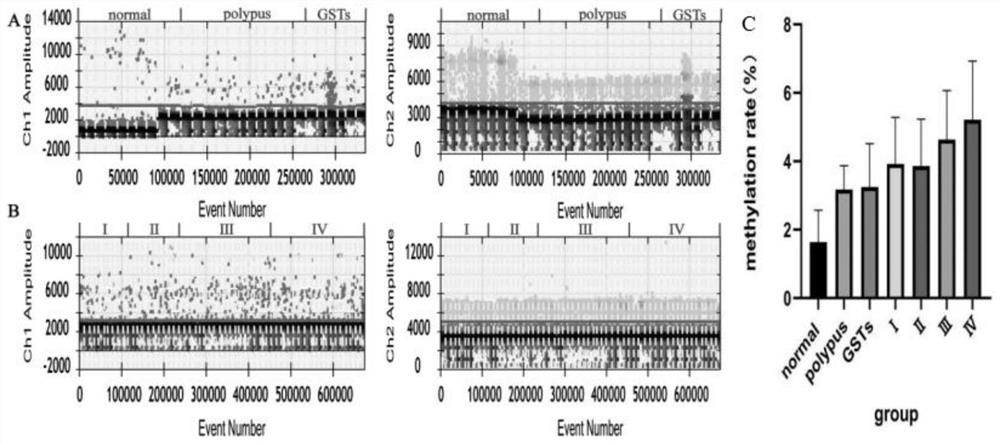
[0079] Design of primers for digital PCR amplification: The methylated sequence and unmethylated sequence of the gene RNF180 after bisulfite conversion were used as target genes to design primers. The primer sequences are listed in Table 4.
[0080] Table 4 Primer sequence list
[0081]
[0082] Among them, the fluorophore labeled at the 5' end of M-Probe is FAM, the quencher group labeled at the 3' end is BHQ1; the fluorophore labeled at the 5' end of U-Probe is VIC, and the quencher group labeled at the 3' end is The killing group is BHQ1.
[0083] 1) Assuming that the sample to be tested is N, the configuration of the reaction system is shown in Table 5:
[0084] Table 5 ddPCR reaction system
[0085]
[0086] 2) After fully mixing the reaction system, dispense it into 17 μL of each reaction tube;
[0087] 3) Take 3 μL of bisulfite-converted DNA and add them to corresponding reaction tubes, with a final volume of 20 μL;
[0088] 4) Put a new droplet generation card...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com