Method for specifically knocking out soybean SOC1 gene by CRISPR/Cas9 and application of method
A SOC1, specific technology, applied in the field of plant genetic engineering and bioengineering, can solve problems such as unclear specific functions, achieve the effect of increasing the number of main stem nodes, increasing the yield of a single plant, and increasing the yield
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
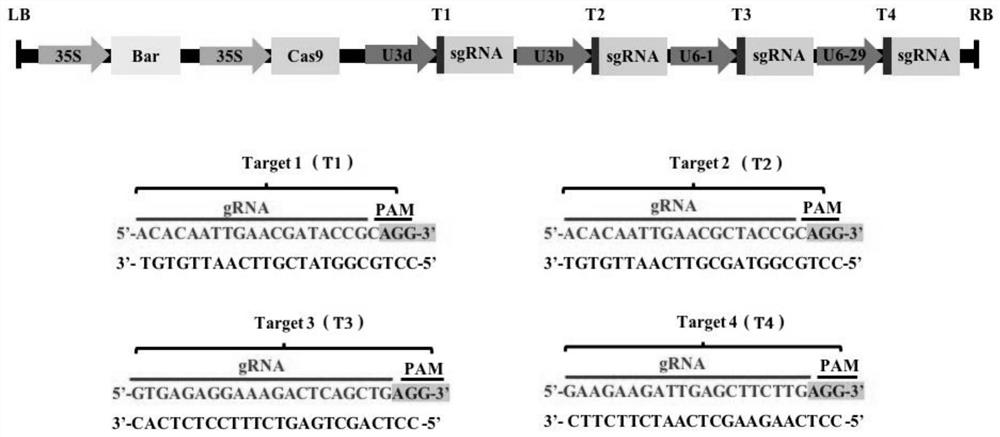
[0031] Example 1 Design and construction of CRISPR / Cas9 vector
[0032] In this example, based on the CRISPR / Cas9 gene editing system, the soybean homologous genes SOC1a and SOC1b were edited, and a CRISPR / Cas9 knockout vector for simultaneously targeting and editing the coding regions of SOC1a and SOC1b was created. The specific creation method is as follows:
[0033] 1. Target primer design
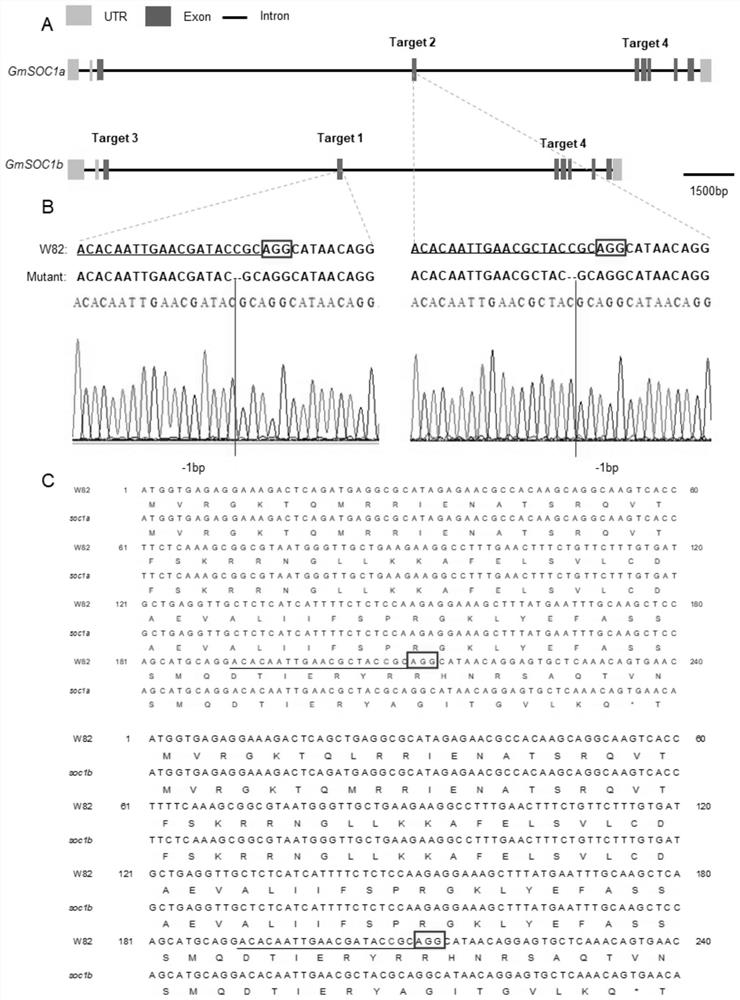
[0034] According to SOC1a provided by NCBI (National center for biotechnology, https: / / www.ncbi.nlm.nih.gov / ) and Phytozome (https: / / phytozome.jgi.doe.gov / pz / portal.html) two databases (Glyma.18G224500) and SOC1b (Glyma.09G266200) gene sequences, using the CRISPR direct website (http: / / crispr.dbcls.jp / ) to design target site adapter primers. Click Specificity check-Soybean(Glycine max)genome,v2.0(Nov, 2015)-design, and select the first 20 bp of the appropriate PAM sequence (NGG) as the target primer, including 1 / 3 / 4 targets located in SOC1b and Located on 2 / 4 targets of SOC1a.
[003...
Embodiment 2
[0051] Example 2 Identification of CRISPR / Cas9 Knockout Vector Genetically Transformed Soybean and Transgenic Soybean
[0052] 1. Stable genetic transformation of soybean with CRISPR / Cas9 knockout vector
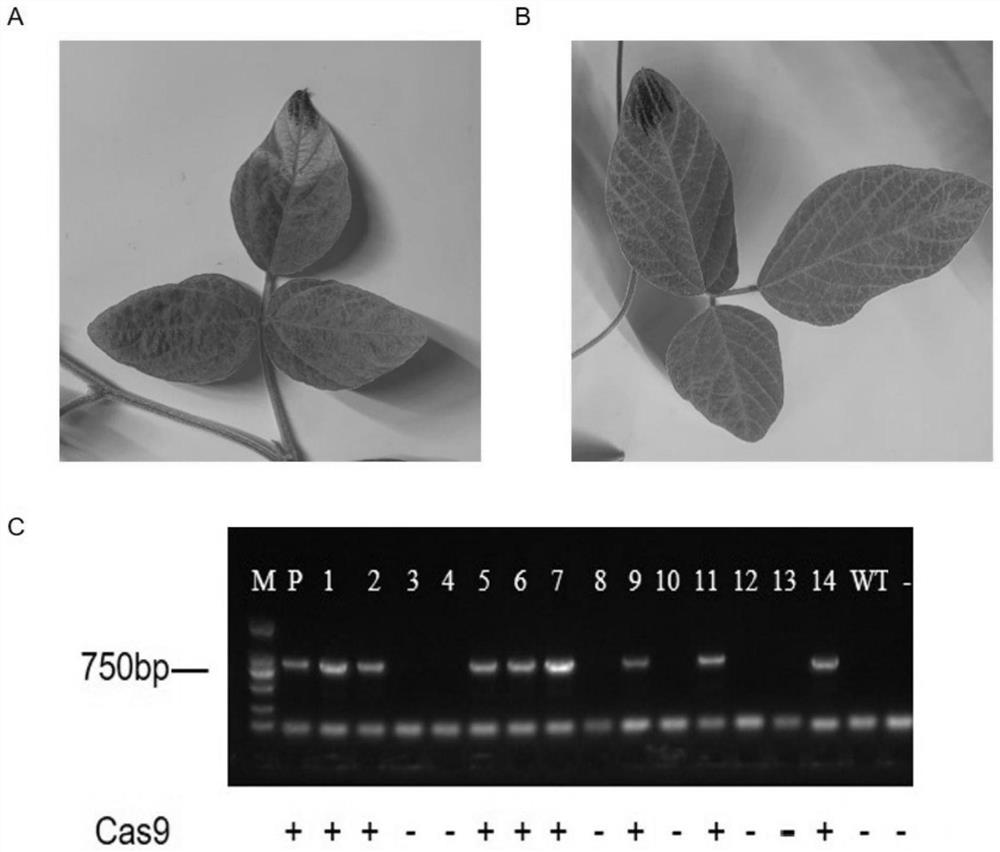
[0053] 1) Hair root transformation and target editing effect detection
[0054] The CRISPR / Cas9 knockout vector constructed in Example 1 containing four tandem target sites was transformed into Agrobacterium K599, cultured at 28°C for 36 hours, and positive clones were selected to infect the cotyledon of Willimas82 soybean to induce hairy roots. The hairy roots grown for 15 days under light culture at 25°C were taken for DNA extraction (CWBIO DNA Kit). Cas9-specific primers (SEQ ID.NO.9 and SEQID.NO.10) were used to detect positive hairy roots, and target editing detection primers (SEQ ID.NO.11~SEQ ID NO.20) were used to perform PCR amplification and analysis of positive hairy roots. Sequencing, to count the editing efficiency of the target. The sequencing results showed ...
Embodiment 3
[0067] Phenotype identification of embodiment 3 soc1a1b double mutant plants
[0068] Homozygous soc1a1b double mutant plants and control Willimas82 (W82) plants were planted in the potted field of the Northeast Institute of Geography and Agroecology, Chinese Academy of Sciences, under natural long-day conditions (16 hours of light / 8 hours of darkness) and artificially set short-days (light 12 hours / dark 12 hours). Observe their flowering and maturity stages, and count the number of nodes and yield per plant when they are mature. The flowering period, maturity period, number of nodes and yield per plant of mutant plants and W82 plants are as follows: Figure 4 shown.
[0069] in, Figure 4 A is the phenotype of flowering stage, maturity stage, number of nodes and yield per plant of W82 and soc1a1b double mutant plants under short-day conditions in 2019; Figure 4 B is the phenotype of flowering stage, maturity stage, number of nodes and yield per plant of W82 and soc1a1b d...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com