Agrobacterium binary expression vector
A binary expression vector, Agrobacterium technology, applied in the direction of introducing foreign genetic material, bacteria, biochemical equipment and methods using vectors, etc., can solve the inability to monitor the expression of the target protein in situ and real-time, and the inability to realize the expression of the target protein. Visualization etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0043] Deletion of the GUS screening marker in the original vector pCambia2301 of embodiment 1
[0044] 1.1 Utilize recombinase Redα / β or RecE / T-mediated homologous recombination to precisely modify DNA molecules to delete GUS screening markers and their promoters and terminators. The steps include: adding the original pCambia2301 vector (Invitrogen Corporation ) was digested at 37° C. with restriction endonucleases SnaBI and MaubI (Thermofisher Company) to obtain a linearized pCambia2301 vector.
[0045] 1.2 Then, the above-mentioned linearized pCambia2301 vector and primers GUS-deletion-F and GUS-deletion-R were recombined in DH10β Escherichia coli (NEB Company) in vivo using Red / ET technology. Recombinant Escherichia coli DH10β was cultured overnight at 37°C in LB medium containing kanamycin sulfate.
[0046] The primer sequences (5'-3') used are:
[0047] GUS-deletion-F:
[0048] CTGGCGTAATAGCGAAGAGGCCCGCACCGATCGCCCTTCCGATAAATTATCGCGCGCGGTGTCATCTATGTTACTAGATC;
[0049]...
Embodiment 2
[0052] Embodiment 2 optimizes the construction of carrier
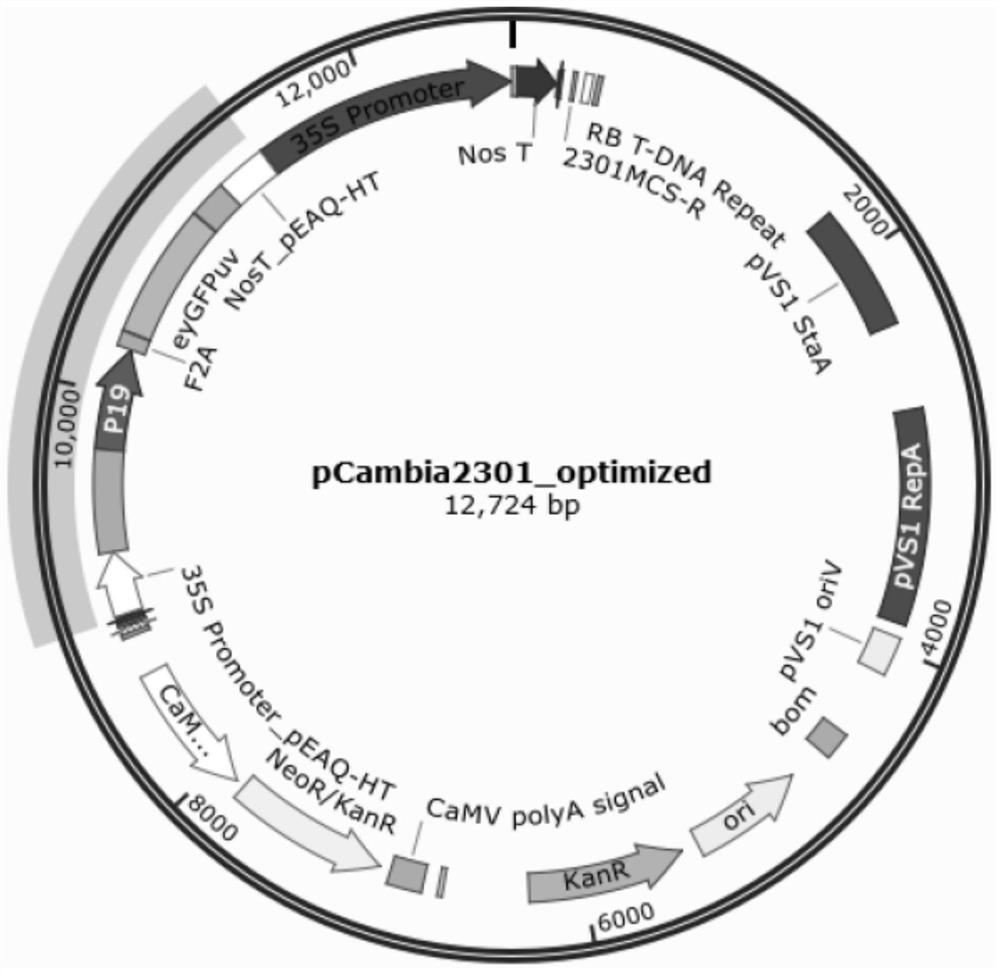
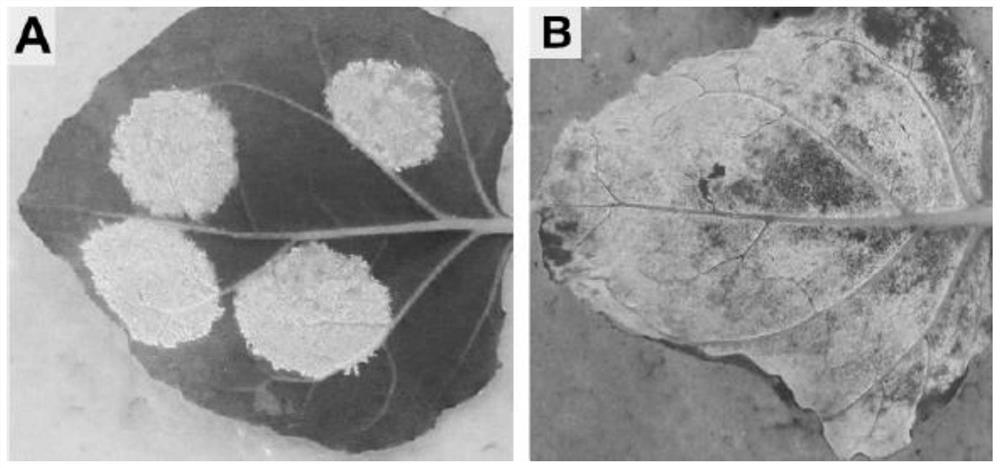
[0053] 2.1 Synthesis of the insert fragment (Insert fragment): The insert fragment contains seven parts: 35S promoter, 5'UTR, P19, F2A peptide, eyGFPuv, 3'UTR and NosT, and the structure is 35S promoter-5'UTR-P19- 2A peptide-eyGFPuv-3'UTR-NosT, the nucleotide sequence of the fusion gene is SEQ ID NO:1.
[0054] The fusion sequence was synthesized by Nanjing GenScript Biotechnology Co., Ltd.
[0055] 2.2 Construction of an optimized vector: Digest the insert fragment synthesized in step 2.1 and the intermediate vector pCambia2301-Gus-Del vector obtained in Example 1 with restriction enzymes ECoRI and SmaI (NEB Company) at 37°C for 12h to obtain Linearized pCambia2301-Gus-Del vector. Then, T4 DNA Ligase (NEB Company) was used to perform ligation according to the instructions of the kit to construct an optimized vector pCambia2301-Optimized, named pF08, which was verified to be correct by sequencing. The nucleotide se...
Embodiment 3
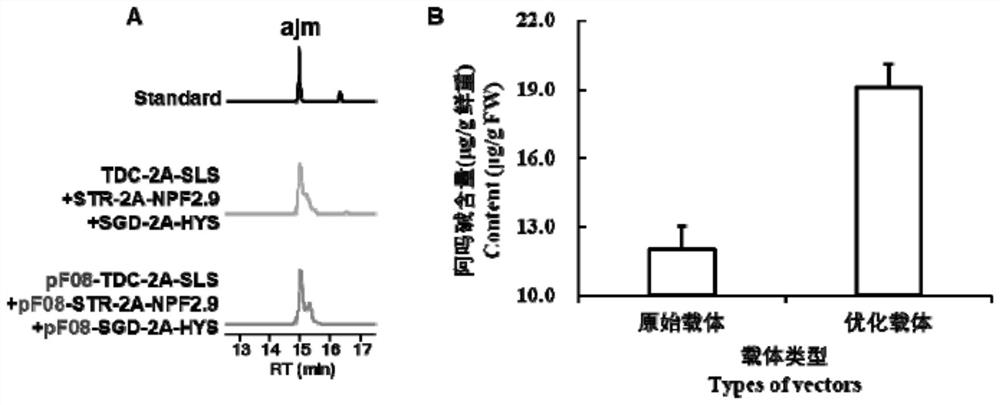
[0056] Example 3 Amplification of Amoline Biosynthesis-Related Genes
[0057] Referring to the method disclosed in the patent document CN202010092362.3, the target genes TDC, SLS1, STR, NPF2.9, SGD and HYS related to amoline biosynthesis were amplified. Including the following steps.
[0058] 3.1 Extraction of periwinkle total RNA and reverse transcription
[0059] The periwinkle leaves were ground into powder with liquid nitrogen, and 100 mg of the ground powder was placed in a centrifuge tube with 1.5 mL of RNA free, and then RNA was extracted according to the instructions of Quanjin’s RNA extraction kit. The extracted RNA is directly reverse-transcribed to produce cDNA.
[0060] Take 1 μg of RNA and reverse transcribe to produce cDNA using Quanshijin reverse transcription kit.
[0061] 3.2 Target gene cloning
[0062] Use the periwinkle cDNA obtained by reverse transcription in step 3.1 as a template, and use the primers shown in Table 1 to amplify according to the foll...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com