Detection method for replicative AAV
A detection method and technology to be detected, applied in the directions of microorganism-based methods, biochemical equipment and methods, and microorganism determination/inspection, etc., can solve the problems of false positives, high requirements on the culture environment, and difficulty in reaching the PCR detection limit, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
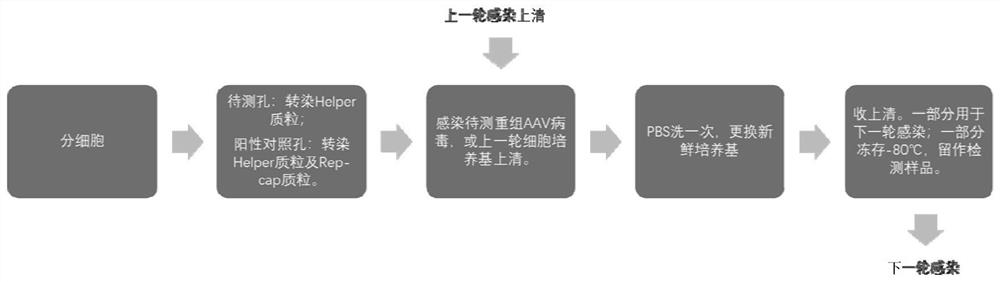
[0032] Example 1: Transfection and infection of HEK293T cells
[0033] The first round of infection experiment process:
[0034] 1.Day1. Digest HEK293T cells 24 hours in advance, count, according to 1×10 per well 6 Cells / 2ml were spread to 6-well plates (pretreated with 100ug / ml poly-lysine-PBS solution for 10min), a total of 4 wells. Another 1×10 6 The cells were plated to a 6cm dish for passage. Incubate overnight at 37°C.
[0035] 2.Day2.4 wells of cells, 2 wells are duplicate wells. Two of the wells were transfected with 2ug pHelper plasmid, and the other two wells were transfected with 1ug pHelper+1ug pRC8 plasmid.
[0036] 3. Day 3. Infect the recombinant AAV virus to be tested.
[0037] 4.Day4. Wash once with PBS and replace with 1.5ml of fresh DMEM-10% FBS medium.
[0038] 5.Day5. Collect the medium supernatant in a 1.5ml EP tube and centrifuge at 1000rpm for 5min at room temperature to remove cells and cell debris. Take 100ul of the supernatant and freeze it a...
Embodiment 2
[0045] Embodiment 2: Cell culture supernatant sample DNase I treatment
[0046] After 4-5 rounds of the infection process described in Example 1, a total of 16 samples to be tested were collected, and the samples were operated as follows:
[0047] 1. Take it out of the -80°C refrigerator and thaw at room temperature.
[0048] 2. Configure DNase I mix in the EP tube, add 640ul nuclease-free water, 160ul 10*DNase IBuffer, 16ul 5U / ul DNase I, and mix by pipetting. Add to two 8-strip PCR tubes in turn.
[0049]3. Add 50ul samples (2-fold diluted samples) sequentially.
[0050] 4. PCR instrument, 37°C for 30min, 75°C for 15min.
[0051] 5. After the treatment, the samples were vortexed and mixed, and then centrifuged briefly.
[0052] 6. Take 10ul sample and add 90ul nuclease-free water (20 times diluted sample), vortex to mix, and centrifuge briefly.
Embodiment 3
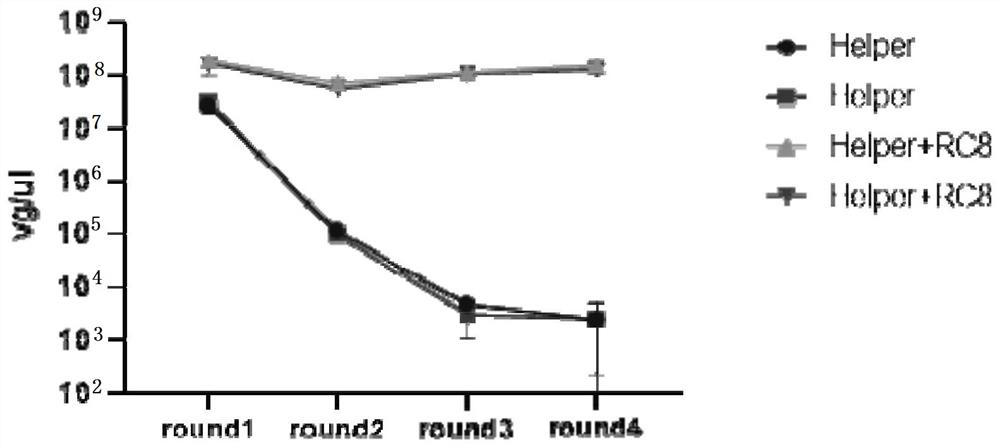
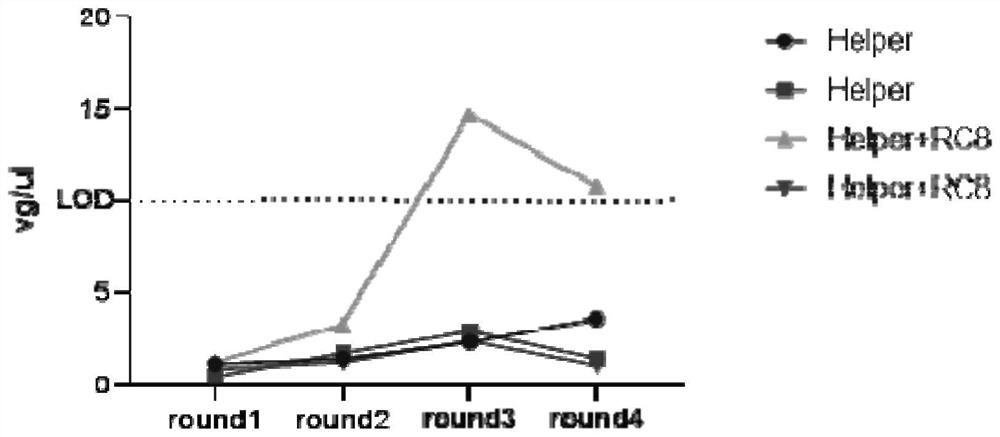
[0053] Example 3: Using taqman qPCR technology to detect rcAAV in the culture supernatant of each round of infected cells
[0054] The processed samples were configured with a PCR system according to the following system:
[0055] 2*AceQ qPCR probe master mix 10ul 50*ROX reference Dye 1 0.4ul Primer F (10uM) 0.4ul Primer R (10uM) 0.4ul Taqman probe(100uM) 0.02ul Template DNA 2ul wxya 2 o
6.8ul Total 20ul
[0056] rcAAV primer SET:
[0057] ITR-P5-taqF AGTGGCCAACTCCATCACTA
[0058] ITR-P5-taqR CCTCTAATACAGGACCTCCCTAAC
[0059] ITR-P5-probe CGTAATTCACGTCACGACTCCACCC
[0060] BGH primer SET:
[0061] BGH-F CCAGCCATCTGTTGTTTGCC
[0062] BGH-R ACTCAGACAATGCGATGCAAT
[0063] BGH-Probe CCCGTGCCTTCCTTGACCCT
[0064] Carry out PCR reaction on ABI7500 according to the following conditions:
[0065]
[0066] Since the positive control wells can support the continuous packaging or continuous replication of recom...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Titer | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com