Construction method of reference protein database, storage medium and electronic equipment
A technology for building methods and databases, applied in proteomics, instrumentation, genomics, etc., which can solve problems such as increasing the cost of experiments and analysis, high false positive results, and biased results.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0075] The preparation of embodiment 1 sample
[0076] Preparation of tryptic peptides
[0077] About 105 K562 cells were lysed by adding 8M urea containing 100mM ammonium bicarbonate for 30 minutes. Cell lysates were then diluted with 600 μL of 100 mM ammonium bicarbonate (reducing urea to 2M) and reduced with DTT (final concentration 10 mM). The protein was alkylated by adding IAA (final concentration 55 mM), and digested by adding 100 mg trypsin. Peptides were desalted using SepPak Plus columns, dried by vacuum centrifugation and diluted with 0.1% acetic acid. Approximately 200 ng of tryptic digested peptide was used for analysis in each experiment.
[0078] The preparation process of the tryptic peptide of the present invention can also be prepared by methods known in the art.
[0079] Perform mass spectrometry analysis on the above tryptic peptides to obtain mass spectrometry data files. The mass spectrometry here can adopt the existing conventional methods and instr...
Embodiment 2
[0080] Example 2 Construction method of GVP reference protein database based on sample
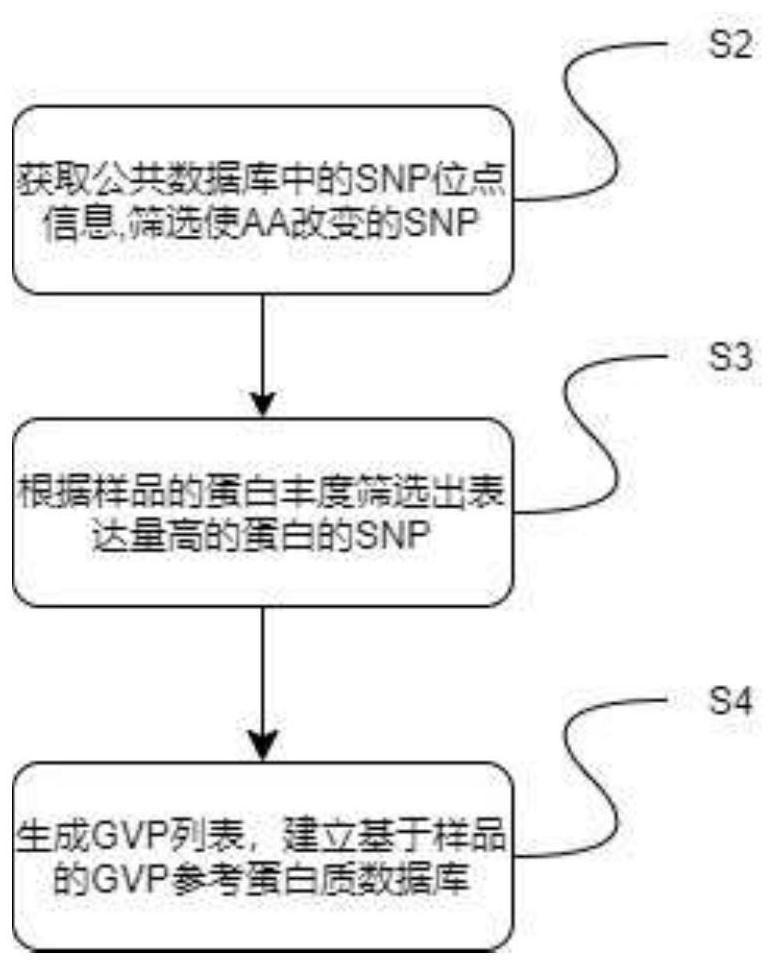
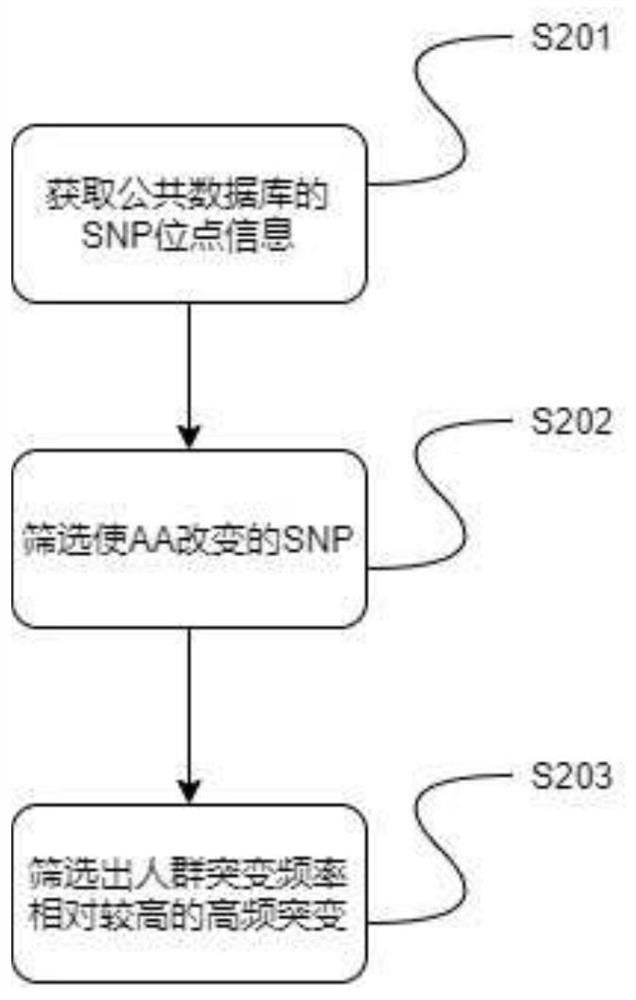
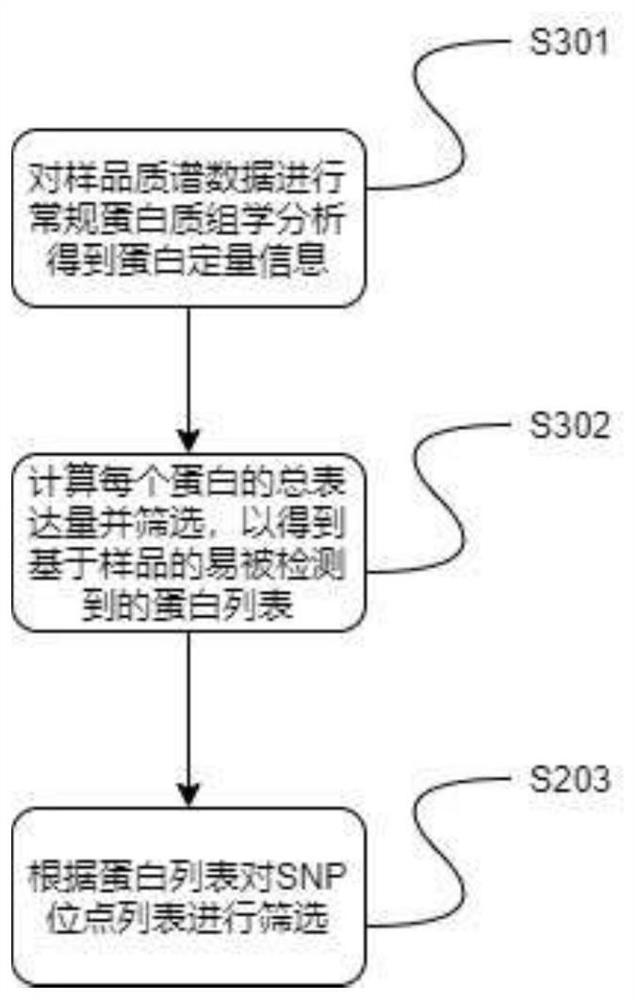
[0081] In this example, the GVP reference protein database was constructed based on the samples in Example 1. Schematic flow chart of the construction method of the GVP reference protein database of Example 2, figure 2 for figure 1 The detailed method flowchart schematic diagram of step S2 in, image 3 for figure 1 The detailed method flow diagram of step S3, Figure 4 for figure 1 The detailed method flow diagram of step S4. Such as figure 1 , figure 2 , image 3 , Figure 4 As shown, the specific steps of this embodiment are as follows:
[0082] First, step S2 is performed to obtain SNP site information in the public database, and screen the SNP sites to obtain a list of SNP sites that cause changes in encoded amino acids.
[0083] Specifically, as in step S201, download the VCF file of all chromosomes from the official website of 1000genome, and download the common_all.vcf....
Embodiment 3
[0123] Example 3 The method for determining SNP based on proteomics
[0124] In this example, the reference protein database and the GVP list obtained by the construction method of the sample-based GVP reference protein database in Example 2 were used to determine the SNP based on proteomics for the samples in Example 1.
[0125] Figure 5 It is a schematic flow chart of the method for determining SNP based on proteomics. Such as Figure 5 As shown, first, as in step S1, protein extraction is performed on the sample, proteomic detection is performed with a mass spectrometer, and mass spectrometry data is collected.
[0126] Then construct the GVP reference protein database and the GVP list based on the sample as steps S2 to S4, including steps:
[0127] Step S2, obtaining SNP site information in the public database, screening the SNP sites, and obtaining a list of SNP sites that cause changes in encoded amino acids.
[0128] Step S3, in the SNP site list, according to the ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com