Close linkage marker for controlling related genes of soybean leaf color and application of close linkage marker
A soybean leaf and gene technology, applied in the field of breeding and molecular genetics, can solve problems such as low efficiency and restricting the development of soybean functional genomics
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0028] 1. For the phenotypic identification of wild-type and mutant materials, refer to "Description Specifications and Data Standards for Soybean Germplasm Resources" edited by Qiu Lijuan et al. From 2016 to 2018, at the Beijing Shunyi Experimental Station of the Institute of Crop Science, Chinese Academy of Agricultural Sciences, the leaf yellow mutant H745 and the wild-type soybean variety ZP661 were observed and identified for the entire growth period, especially the field phenotypes of the blooming and mature stages. Chlorophyll content identification was carried out at the full flowering stage, and 10 plants of ZP661 and H745 were selected for testing at the mature stage, and the main agronomic traits such as plant height, bottom pod height, number of nodes, number of branches, number of pods per plant, and 100-grain weight were measured and photographed. .
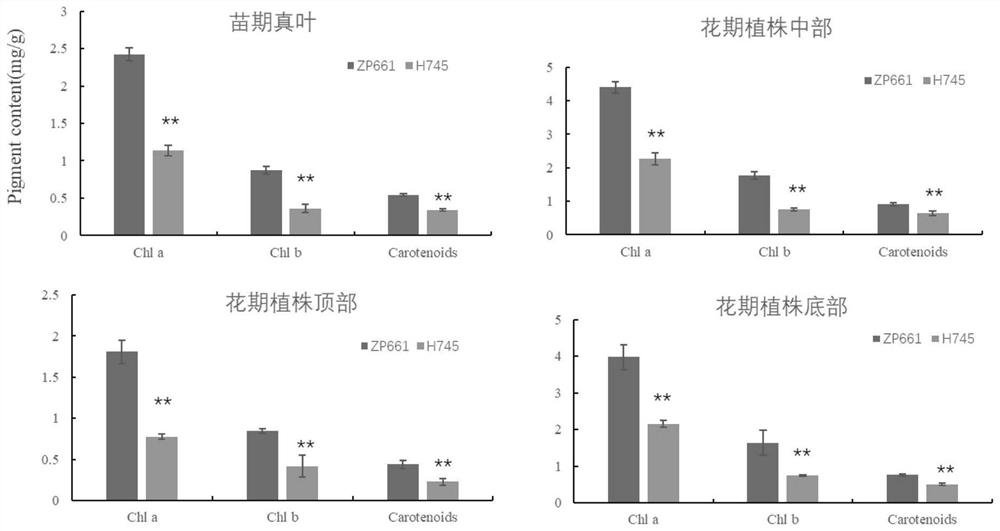
[0029] 2. Determination of photosynthetic pigment content
[0030] At the full flowering stage (R2), the wild ty...
Embodiment 2
[0040] 1. Genetic population construction
[0041] At the Beijing Shunyi Experimental Base of the Chinese Institute of Agricultural Sciences, ZP661 was used as the female parent and the yellow mutant H745 was used as the male parent to prepare a hybrid combination and obtain hybrid seeds. In the summer of 2017, the F1 generation plants were propagated and the seeds were harvested at the same location. In 2018, the F2 generation population was planted at the same location, and the F3 generation population was continued to be planted and phenotyped in 2019 to verify the genotype of the F2 population.
[0042] 2. Genomic DNA Extraction
[0043] Sample preparation: Weigh 0.1g to 0.5g of young soybean leaves and place them in a 2mL centrifuge tube, mark the sample number on the tube cover and tube wall, add steel balls, freeze in liquid nitrogen, and shake on a shaker until Grind the leaves into powder. Cleavage: Add 2% CTAB (1 / 1000 volume β-mercaptoethanol) extract at 600°C, sha...
Embodiment 3
[0065] Based on the sequencing data in the candidate interval, 9 SNP sites located on the gene were selected from the 19 SNP sites in the association interval that conformed to the rule of BSA pool construction ( Figure 8), respectively for SNP verification and CAPS / dCAPS development (according to the variant sites obtained in the resequencing data, and then obtain the SNP positions from the Phytozome website https: / / phytozome.jgi.doe.gov / pz / portal.html# Click on the relevant sequence in the attachment to design primers for identification). After identification, the SNP at H745-1 among the 9 sites was false, the amplified fragment of H745-4 was a homologous sequence, and the amplified band of H745-8 was double peaked; the other 6 sites were all mutant SNPs (i.e. The variation is consistent with the sequencing results), successfully developed as CAPS / dCAPS markers for identification of populations.
[0066] 481 F2:3 populations were detected by 6 CAPS / dCAPS markers, and the c...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com