Fluorescent biosensor for detecting uracil DNA glycosylase as well as detection method and application of fluorescent biosensor
A biosensor, glycosylase technology, applied in the field of detection and analysis, can solve the problems of limited amplification ability, limited sensitive detection of low-activity UDG, etc., to achieve good precision and reproducibility, good anti-interference ability, high sensitivity Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
specific Embodiment approach
[0055] In yet another specific embodiment of the present invention, the fluorescent biosensor comprises the following DNA sequence:
[0056] Walking chain 1:
[0057] Track chain H1:
[0058] Track chain H2:
[0059] Lock chain: ACCGGCUCGGAGTT (SEQ ID NO. 4).
[0060] Wherein, the italic parts of orbital chains H1 and H2 represent complementary sequences. The underlined parts of orbital chains H1 and H2 indicate the binding sites of the corresponding walking chains. rA in orbital chains H1 and H2 represents adenine ribonucleic acid; the bold part of walking chain 1 and orbital chain H1 represents the catalytic core of 8-17DNAzyme; "FAM" represents 6-carboxyfluorescein.
[0061] In yet another specific embodiment of the present invention, a method for detecting uracil DNA glycosylase is provided, the method comprising:
[0062] 1) Incubate the reaction with the analyte, endonuclease IV and DNA walker 1 to unblock the walking chain;
[0063] 2) Add MgCl to step...
Embodiment 1
[0088] Example 1: Detection of uracil-DNA glycosylase activity based on DNAzyme-driven cascade DNA walker signal amplification strategy
[0089] The specific method is as follows:
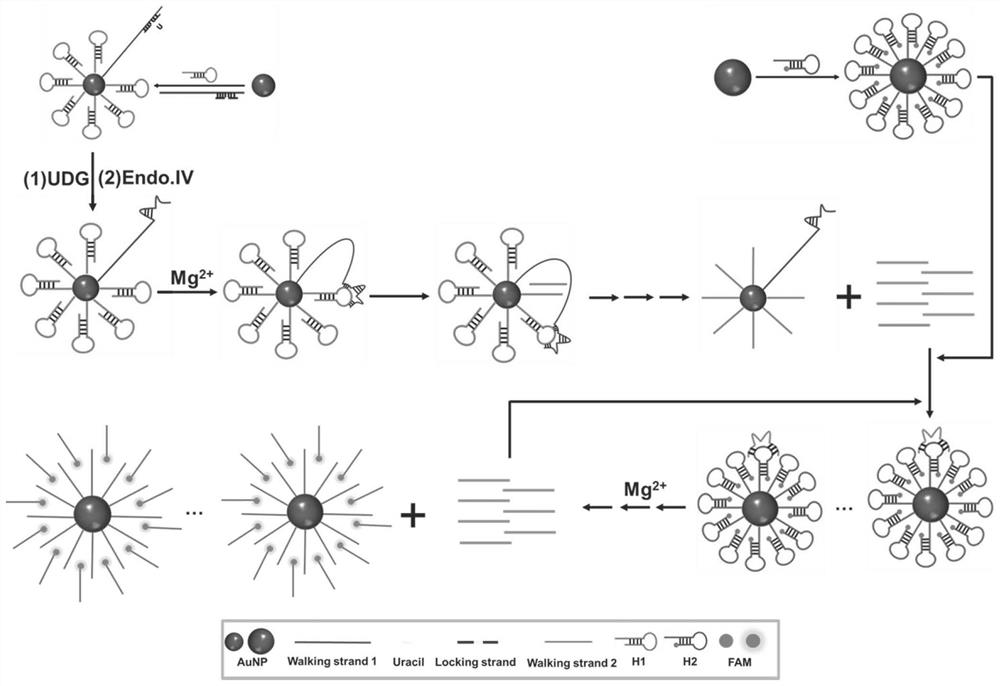
[0090] Mix the locked chain and the thiolated walking chain 1 in 1×Tris-HCl buffer at a molar ratio of 1.2:1, heat at 95°C for 10 min, and then slowly cool to room temperature to form a closed walking chain 1 . The thiolated orbital chains H1, H2 anneal to form a hairpin structure. 13nm gold nanoparticles, closed walking strand 1 and orbital strand H1 were used to construct DNA walker1; 25nm gold nanoparticles and orbital strand H2 were used to construct DNA walker 2.
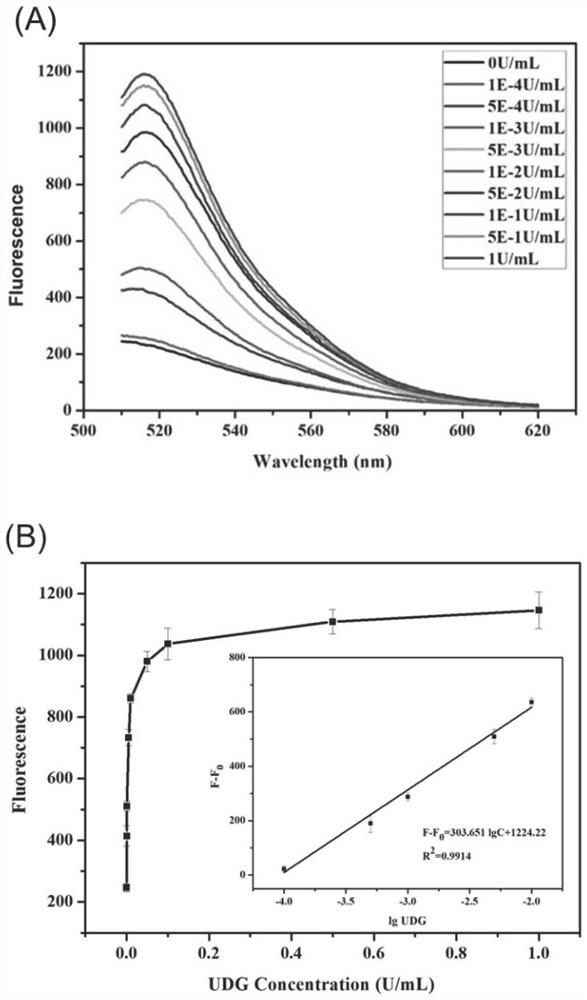
[0091] Different concentrations of UDG, 20U / mL Endo.IV and DNA walker 1 (0.4nM) were incubated in Tris-HCl buffer solution (20mM Tris, 100mM NaCl, pH 8.0) at 37°C for 1h to unblock the walking strand. Then add MgCl 2 (5mM), trigger the walking movement of the primary DNA walker, and incubate at 37°C for 1.5h. Next, add DNA wal...
Embodiment 2
[0092] Embodiment 2: Feasibility study of detection method of the present invention
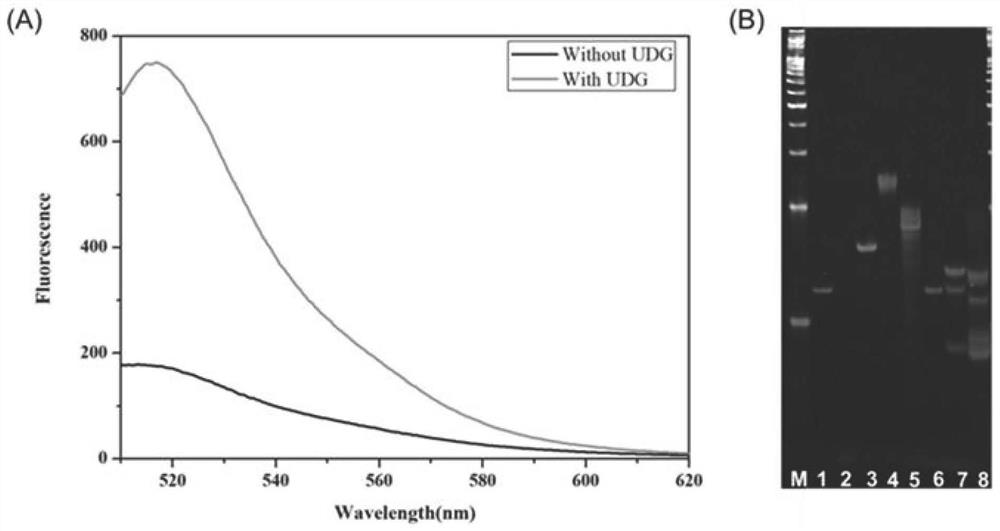
[0093] To verify the feasibility of the proposed DNAzyme-driven cascade DNA walker signal amplification strategy, we first characterized it by fluorescence experiments. As shown in Figure 2A, when there is no target UDG, the fluorescence signal is low, indicating that the DNAzyme is in a closed state, and DNA walker cannot occur. When the target UDG exists, the fluorescent signal is significantly enhanced, indicating that UDG can interact with the locked chain and activate the DNAzyme activity of walking chain 1, start the operation of DNA walker 1, generate a large number of walking chain 2 containing DNAzyme activity, and then activate DNA The operation of walker 2 produces a large number of fluorescent signals.
[0094] Then, the feasibility of the strategy was further verified by PAGE experiments. like figure 2 As shown in B, swimming lane 1, swimming lane 2, swimming lane 4 and swimm...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com