Nested fluorescent quantitative PCR (Polymerase Chain Reaction) detection method for Lyme disease spirochetes
A Lyme disease helix and detection reagent technology, which is applied in the field of molecular biology and microbial detection, can solve the problems of low sensitivity and cannot be used to determine the type of infected strains in positive specimens, so as to improve sensitivity and specificity Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0036] Example 1 Design and optimization of nested fluorescent quantitative PCR primers
[0037] Borrelia Lyme disease has a unique rRNA gene structure, that is, there are two copies of 23S rRNA (rrf) and 5S rRNA (rrl) genes, which are repeated in sequence: -rrf-rrl-rrf-rrl-, forming rrf-rrl in the middle Intergenic region, this structure is specific to Borrelia burgdorferi.
[0038] The rrf-rrl site of Borrelia Lyme disease, which can be identified by single point typing, was selected as the target gene. In the first round, the outer round primers of nested PCR were used to enrich the DNA of Lyme disease spirochetes in the samples; in the second round, specific fluorescent quantitative PCR was used for the specific detection of Lyme disease spirochetes.
[0039] First round: common PCR
[0040] The primers are as follows (SEQ ID NO:1-2):
[0041] Upstream primer F1: 5'-CGACCTTCTTCGCCTTAAAGC-3'
[0042] Downstream primer R1: 5'-TAAGCTGACTAATACTAATTACCC-3'
[0043] The sec...
Embodiment 2
[0050] Example 2 Establishment of a nested fluorescent quantitative PCR detection method for Lyme spirochete
[0051] The nested fluorescent quantitative PCR detection method of Lyme disease spirochete provided by the invention comprises the following steps:
[0052] 1. Extract the DNA in the sample;
[0053] 2. Using the DNA extracted in step 1 as a template, use the first round of PCR primers (SEQ ID NO: 1-2) to carry out PCR amplification reaction;
[0054] 3. Using the amplified product in step 2 as a template, use the second round of PCR primers and probes (SEQ ID NO: 3-5) to carry out PCR amplification reaction;
[0055] 4. Analyze PCR products.
[0056] In step 2, the PCR reaction system is: 10 μL of reaction mixture, 0.5 μL of 10 μM upstream and downstream primers, 4 μL of DNA template, and 5 μL of deionized water.
[0057] The PCR reaction program is: 94°C for 2min, 55°C for 1min, 72°C for 2min, 1 cycle; 94°C for 45s, 55°C for 45s, 72°C for 45s, 33 cycles; 94°C for...
Embodiment 3
[0060] Example 3 Specificity investigation of nested fluorescent quantitative PCR detection method
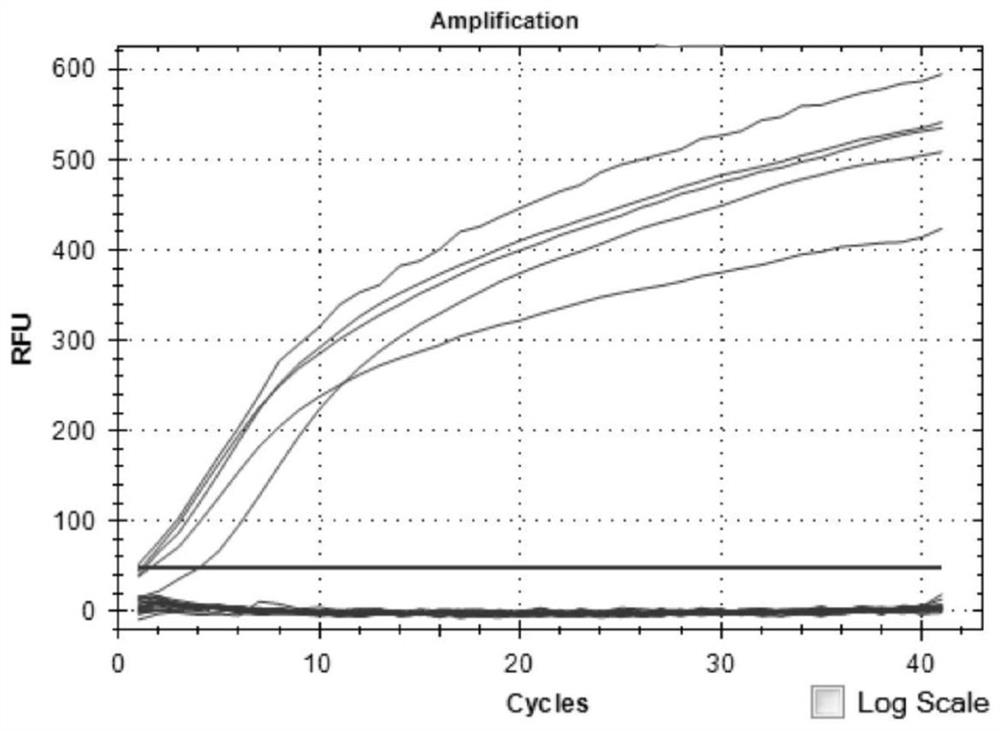
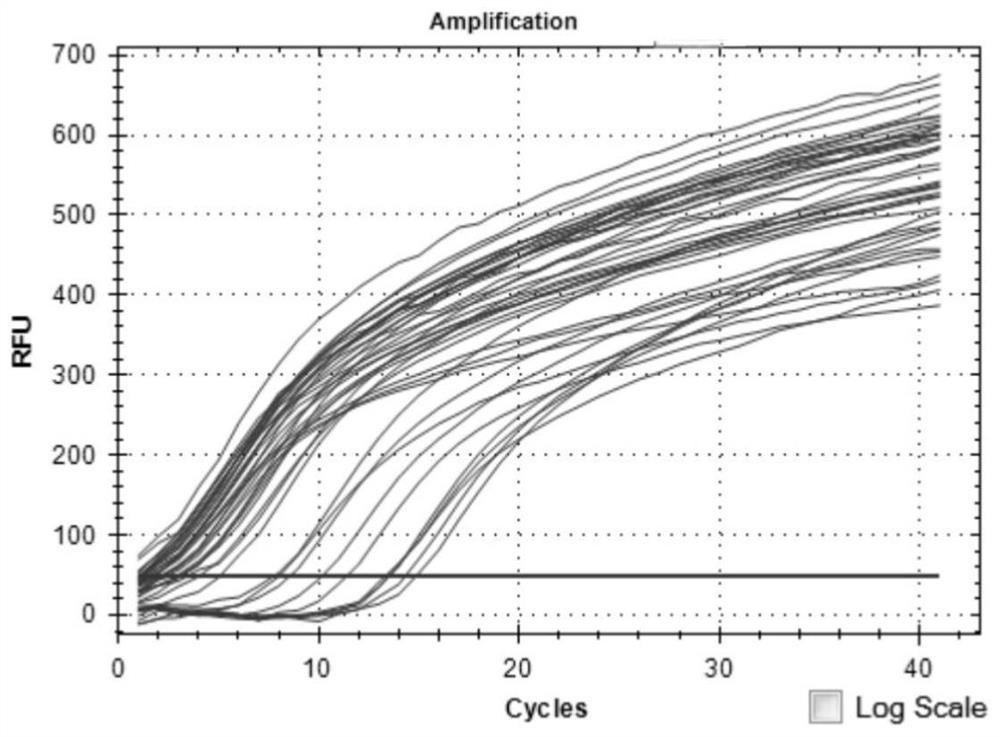
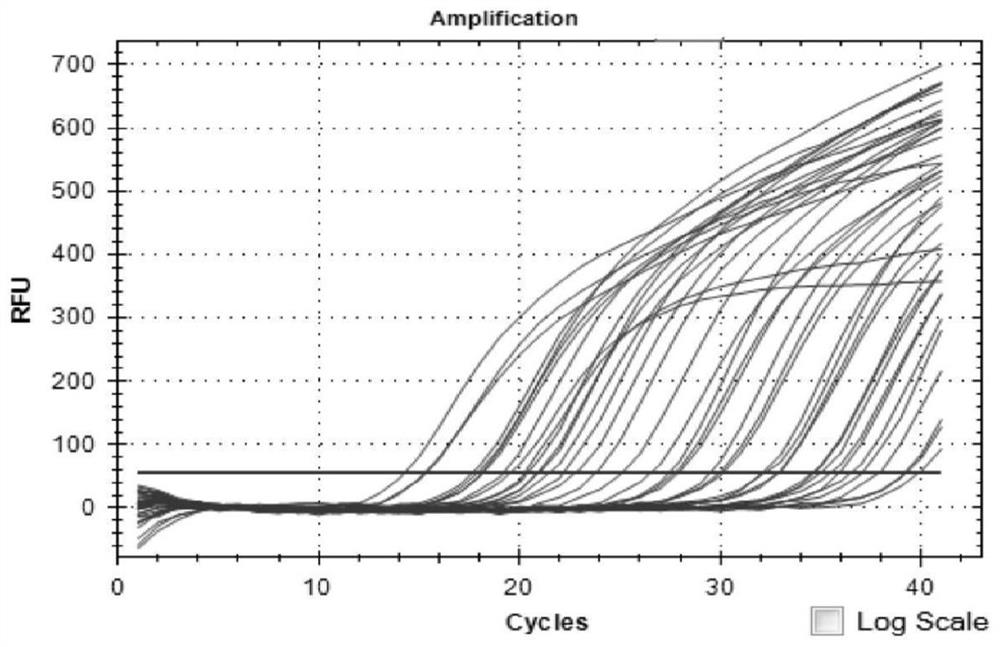
[0061] The strains used included 22 samples of Escherichia coli (1 strain), Rickettsia (5 strains), Bartonella (1 strain), Leptospira (10 strains) and Borrelia burgdorferi (5 strains). The nested fluorescent quantitative PCR method in Example 2 was used for detection. The test results showed that, except Borrelia burgdorferi, other bacteria were not detected, indicating that the method had better specificity ( figure 1 ).
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com