Characterization and inactivation of endogenous retroviruses in chinese hamster ovary cells
A cell and cell line technology that can be used in retroRNA viruses, viruses, viral peptides, etc., to solve problems such as hindering ERV inactivation strategies and lack of clear linkage between genome C-type ERV sequences
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
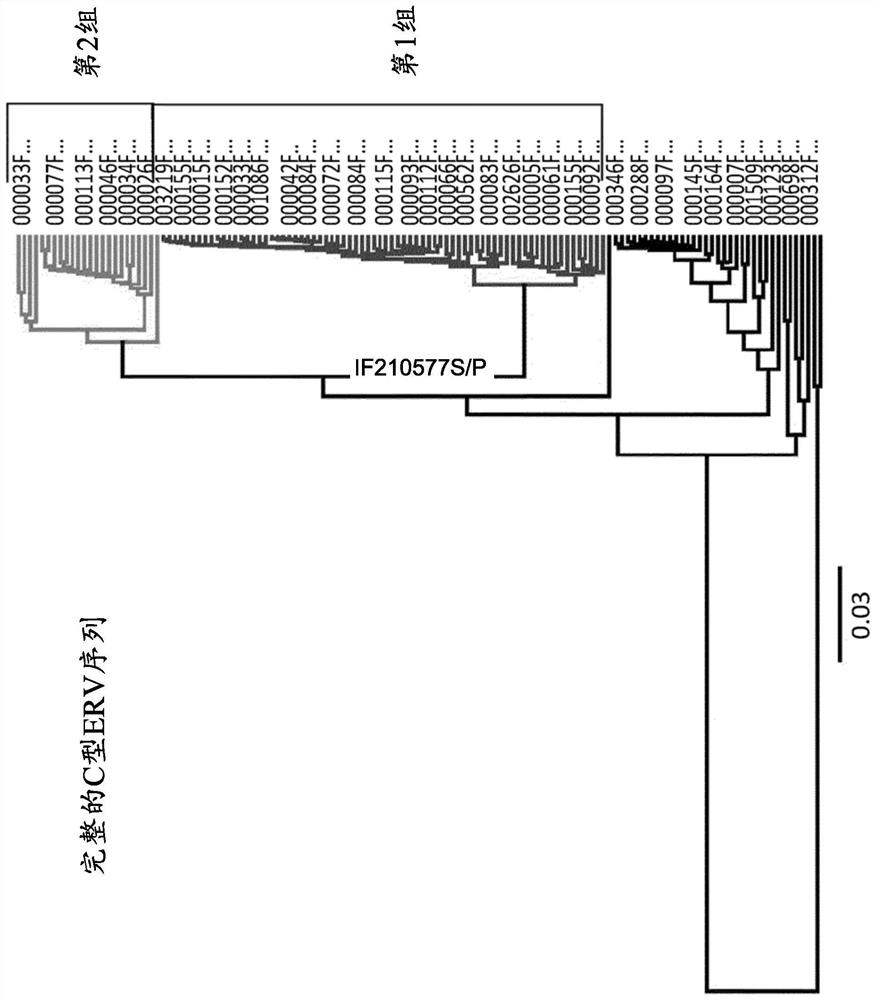
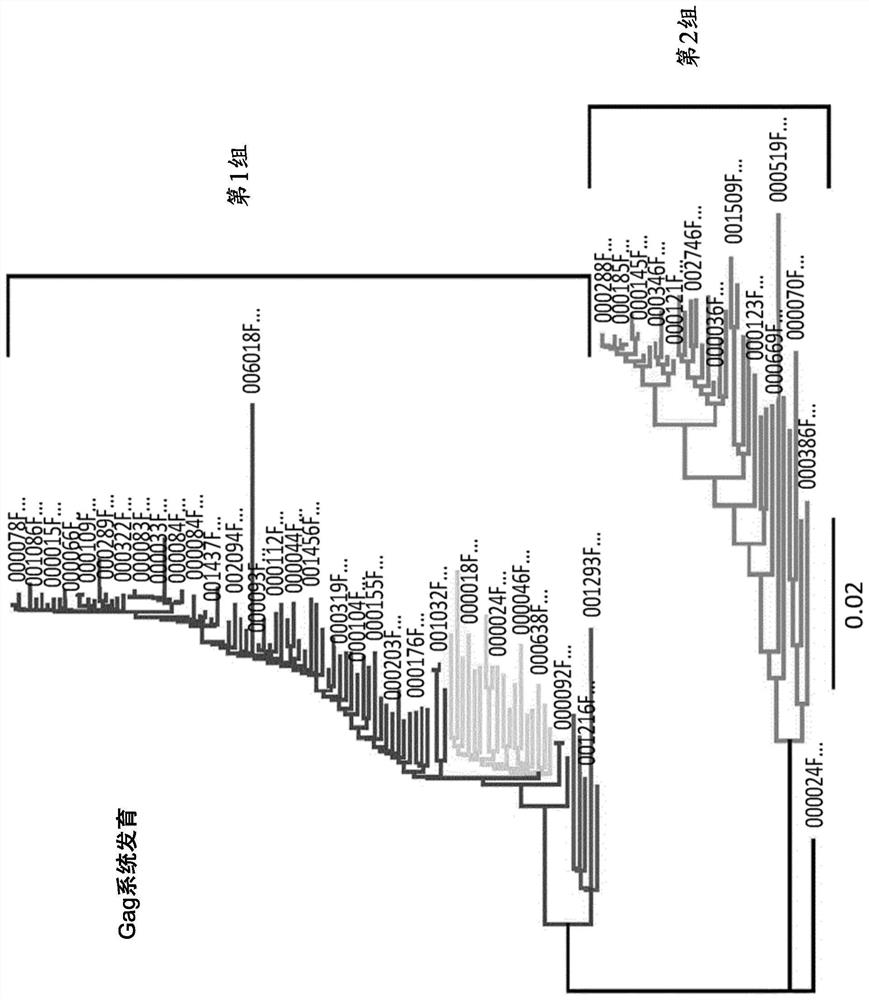
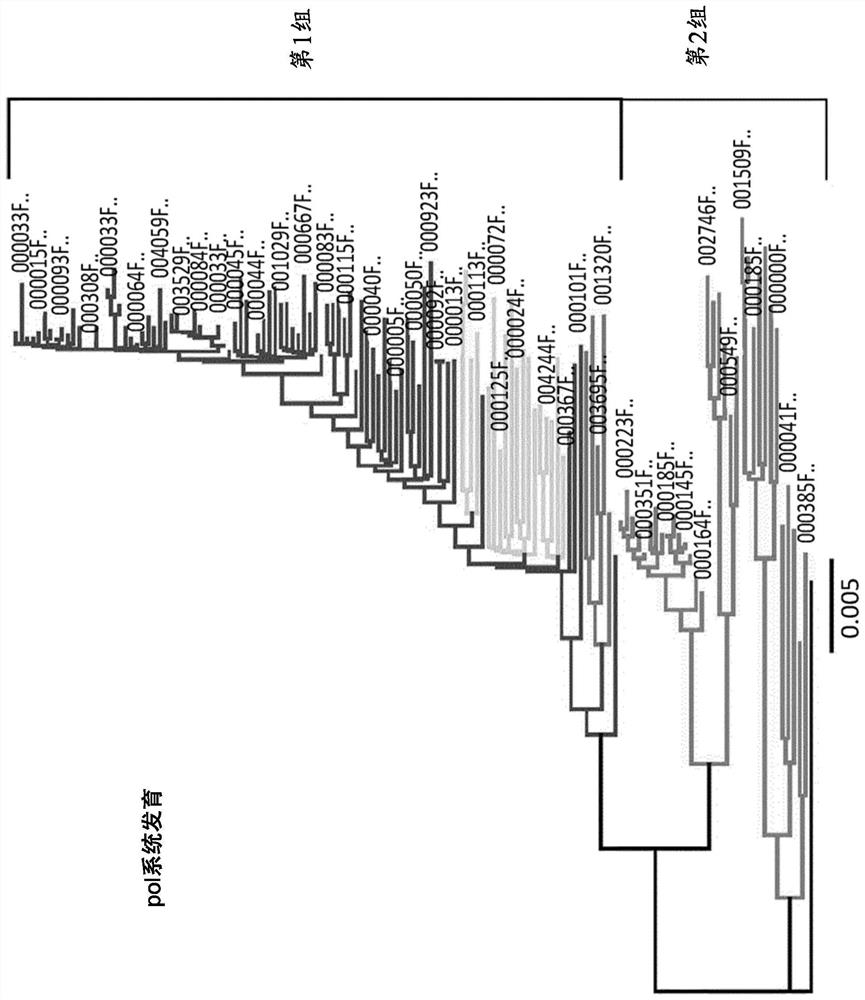
Image
Examples
Embodiment Construction
[0070] Cells according to the invention, preferably mammalian / eukaryotic cells, including engineered cells, can be maintained under cell culture conditions. Standard cell culture conditions are 30°C to 40°C, preferably at or around 37°C, for example in fully synthetic media used for the production of recombinant proteins. Non-limiting examples of this type of cells are non-primate eukaryotic cells such as Chinese Hamster Ovary (CHO) cells, including CHO-K1 (ATCC CCL 61), DG44 and CHO-S cells and SURECHO-M cells (CHO -K1 derivative cells), and small hamster kidney cells (BHK, ATCC CCL 10). Primate eukaryotic host cells include, for example, human cervical carcinoma cells (HELA, ATCC CCL 2) and 293 [ATCC CRL 1573] and 3T3 [ATCC CCL 163] and the monkey kidney CV1 line [ATCC CCL 70], also transformed with SV40 ( COS-7, ATCC CRL1587). The term "engineered" means that the genome of a cell has been altered, for example by insertions, deletions and / or substitutions. Those skilled i...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com