Carboxylic acid reductase mutant with improved catalytic activity and coding gene, genetically engineered bacterium and application thereof
A technology of improving catalytic activity and encoding genes, which is applied in the field of protein engineering, can solve the problems of low catalytic activity and low yield rate, and achieve the effect of high-efficiency biocatalysis and activity improvement
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
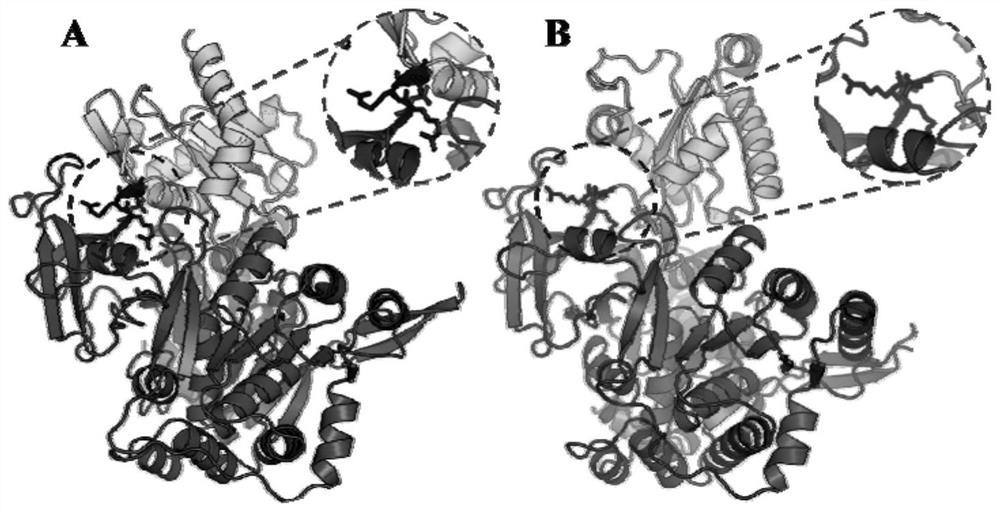
[0026] Example 1 Substrate docking and molecular dynamics simulation
[0027] In order to obtain the spatial structure of the adenylation domain of carboxylic acid reductase MsCAR, homology modeling was performed on the domain. Modeling was performed using the SWISS-MODEL online server (http: / / www.swissmodel.expasy.org / ), and the 3D model of the adenylation state of the domain was based on the carboxylic acid reductase (PDB number: 5MSC) from Nocardia iowensis The structure was obtained as a template, and the 3D model of the thioesterification state was obtained using the structure of carboxylic acid reductase from Mycobacterium marinum (PDB code: 5MSS) as a template. The structure-optimized vanillyl-AMP complex was then docked into the structure of the adenylation state of the MsCAR adenylation domain. The docking results of AMP in the carboxylic acid reductase crystal structure 5MSC were screened, and the best docking posture was selected in combination with the docking s...
Embodiment 2
[0030] Example 2 Virtual mutation of the hinge region of carboxylic acid reductase MsCAR and its screening
[0031] Based on the structure of the adenylation state of the adenylation domain of carboxylic acid reductase MsCAR and the structure of the vanillyl-AMP complex, the point mutation of the protein was carried out in the Build and EditProtein program under the Macromolecules module in the Discovery Studio 2020 software, namely Select the site to be mutated in the structure, select the target amino acid in the Choose AminoAcid operation column to complete the amino acid mutation, and then call the Minimize and Refine Protein program under this module to optimize the structure of the mutated protein. After the optimization is completed That is to say, a more reasonable protein-vanillyl-AMP complex binding conformation has been obtained. Then call the interatomic distance measurement function in the Discovery Studio 2020 software to calculate the distance between the side...
Embodiment 3
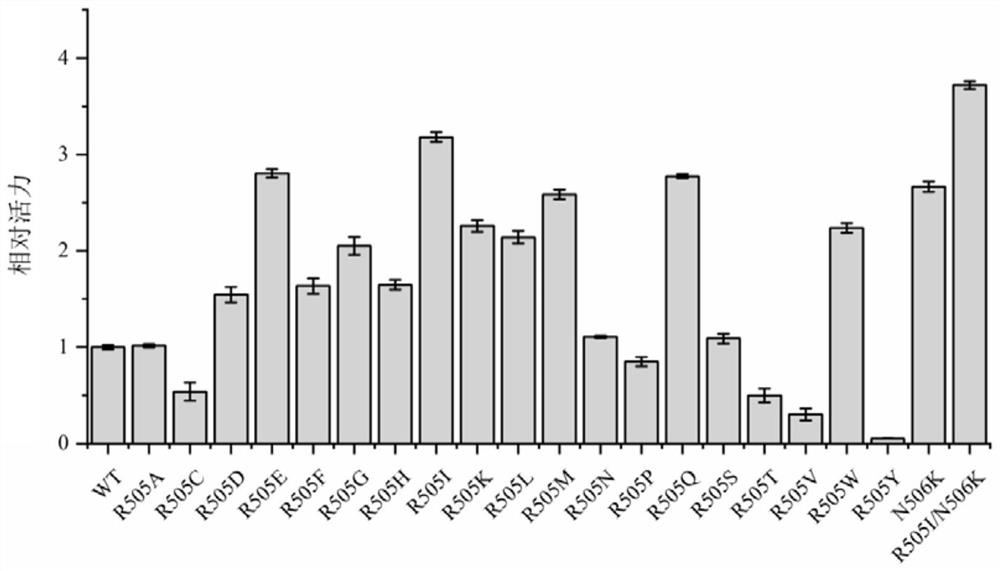
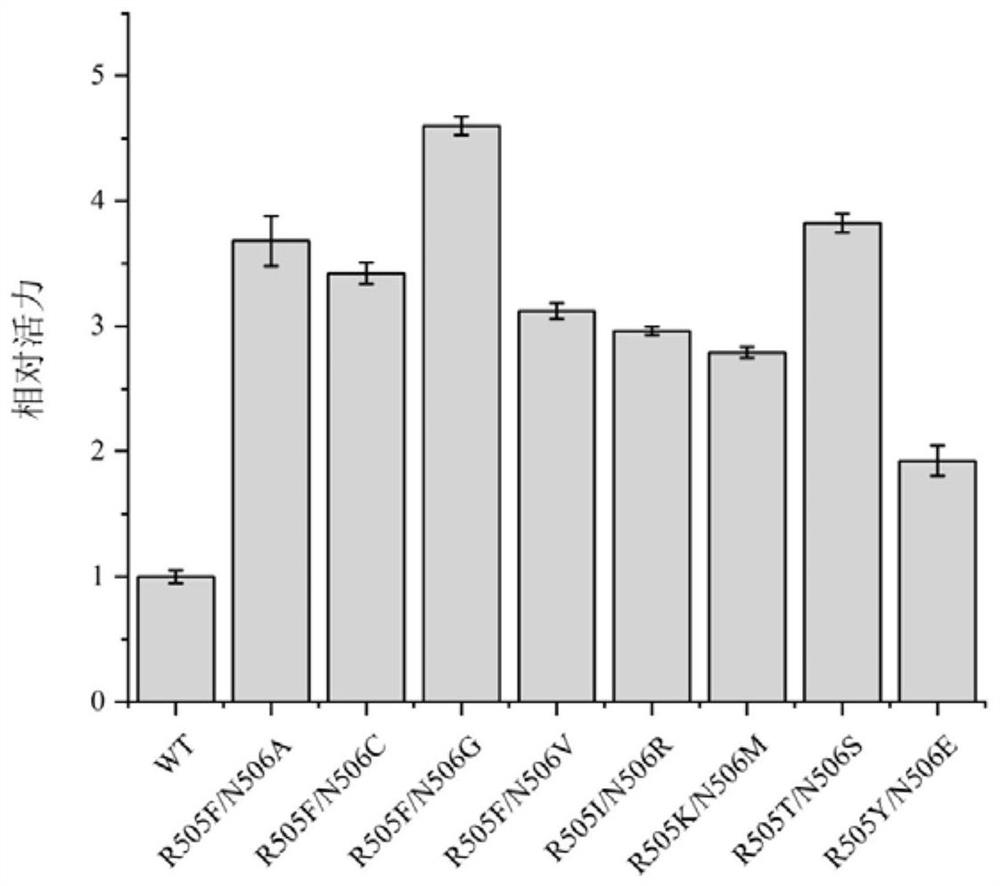
[0033] Construction and screening of embodiment 3MsCAR mutants
[0034] The carboxylic acid reductase MsCAR producing strain was cultured in a large test tube for 10-12 hours, and its plasmid was extracted as a template for subsequent PCR. The KOD-Plus point mutation kit of Japan Toyobo Co., Ltd. was used to construct the mutation library. The specific operation is as follows: 1. According to the product manual, use the target sequence as a template to design primers; 2. Use high-fidelity KOD–Plus-enzyme to introduce point mutations by reverse PCR; 3. Use Dpn I to template plasmid DNA. Digest; 4. Use T4Polynucleotide Kinase and Ligation high in the kit to self-circularize the PCR product; 5. Transform and introduce the resulting cyclized product into E. coli BL21 (DE3) competent cells; 6. Pick a single colony And inoculated in 5mL LB medium, cultivated overnight at 37°C and then sequenced the strains; 7. After ensuring the sequence was correct, each mutant was induced to ex...
PUM
| Property | Measurement | Unit |
|---|---|---|
| catalytic efficiency | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com