Application of CCND1 in preparation of avian retrovirus production enhancer
A retrovirus and enhancer technology, applied in the field of virology, can solve problems such as accelerated virus evolution and mutation, irrationality, and complex diseases, and achieve the effect of promoting replication and prolonging the S phase time
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
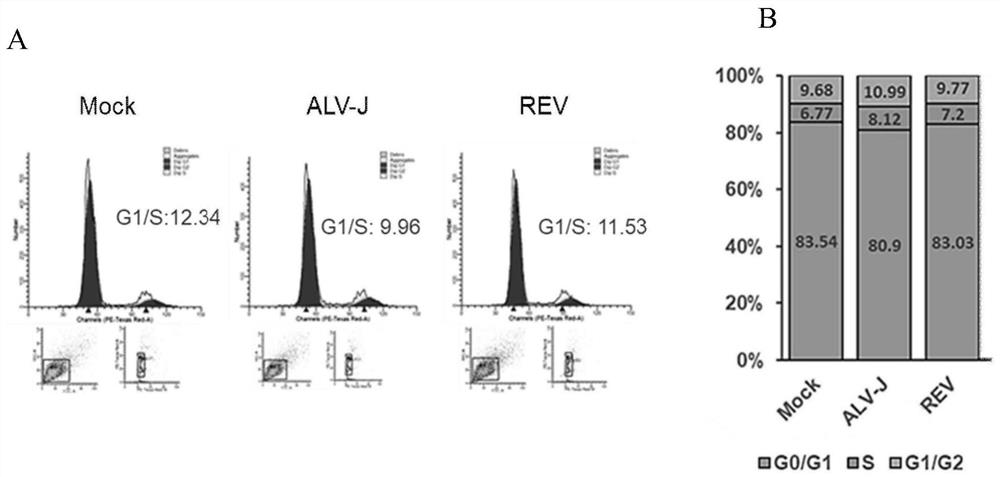
[0036] Example 1 Detection of changes in cell cycle state after avian retrovirus infection of cells
[0037] 1. Cell preparation: DF-1 cells were cultured in 25cm 2 Cell culture flasks, after cell counting, the cell density was adjusted to 0.8×10 8 / well, divided into 3 groups with 3 replicates each:
[0038] In group 1, when the DF-1 cells reached 70% confluence, 1 mL of ALV-J NX0101 virus solution was added, maintained for 2 h, discarded the venom, and added Dulbecco's Modified Eagle Medium (DMEM) medium containing 1% fetal bovine serum, in 5% CO at 37°C 2 The culture was maintained in the incubator for 72 hours, and the cells were collected.
[0039] Group 2, when the DF-1 cells reached 70% confluence, add 1mL REV SNV virus solution, maintain for 2h, discard the venom, add Dulbecco's Modified Eagle Medium (DMEM) medium containing 1% fetal bovine serum, at 37°C 5% CO 2 The culture was maintained in the incubator for 72 hours, and the cells were collected.
[0040] Grou...
Embodiment 2
[0045] Example 2 Proteomic analysis of DF-1 cells infected with ALV-J and DF-1 cells infected with REV
[0046] 1. Cell preparation: add the same cell sample of the same group in Example 1 to the cell lysate RIPA:PMSF (100:1), and harvest the cell protein.
[0047] 2. Protein extraction: After the cells were digested with trypsin, 4 times the volume of lysis buffer (8M urea, 1% protease inhibitor, 3 μM TSA, 50 mM NAM and 2 mM EDTA) were added respectively, and ultrasonically lysed. Centrifuge at 13400×g for 10 min at 4°C to remove cell debris, transfer the supernatant to a new centrifuge tube, and use the BCA kit for protein concentration determination.
[0048] 3. Trypsin hydrolysis: add dithiothreitol to the protein solution to make the final concentration 5mM, and reduce at 56°C for 30min. Afterwards, iodoacetamide was added to make the final concentration 11 mM, and incubated at room temperature in the dark for 15 min. Finally the urea concentration of the sample was dil...
Embodiment 3
[0057] Example 3 Overexpression of CCND1 significantly increases the viral load of ALV-J and REV
[0058] In order to clarify that the activation of CCND1 can promote the replication of ALV-J and REV, this example constructed a CCND1 overexpression plasmid, and verified the promotion effect of CCND1 on the load of ALV-J and REV by overexpressing CCND1.
[0059] 1. Construction of CCND1 overexpression plasmid.
[0060](1) Construction of eukaryotic expression vector. The CCND1 gene is optimized, and the optimized nucleotide sequence is shown in SEQID NO.2. The construction of the eukaryotic expression vector is completed by Gemma Gene Company using the existing technology. The present invention selects pcDNA3.1 eukaryotic expression plasmid, and the Sequence the complete CCND1 gene sequence without mutation and insert it into the plasmid, and then transform the constructed plasmid into the competent cell DH5α. After the transformation is completed, shake the bacteria to extrac...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com