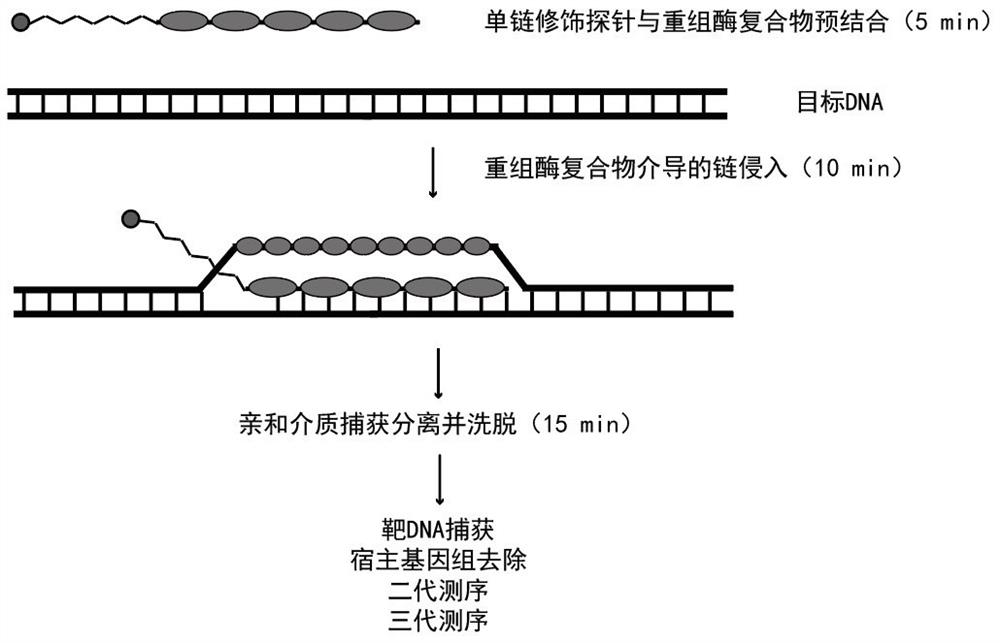
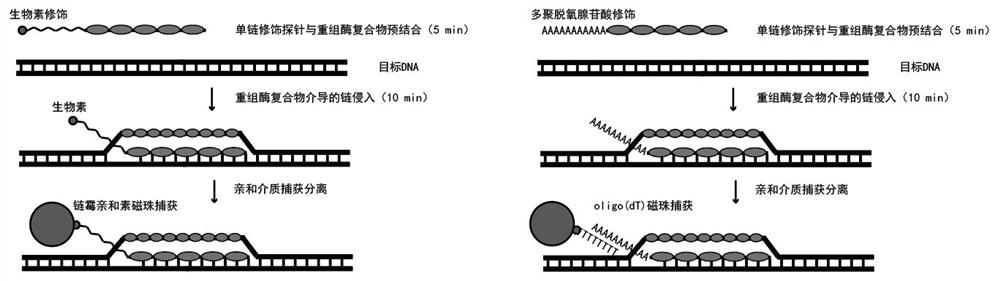
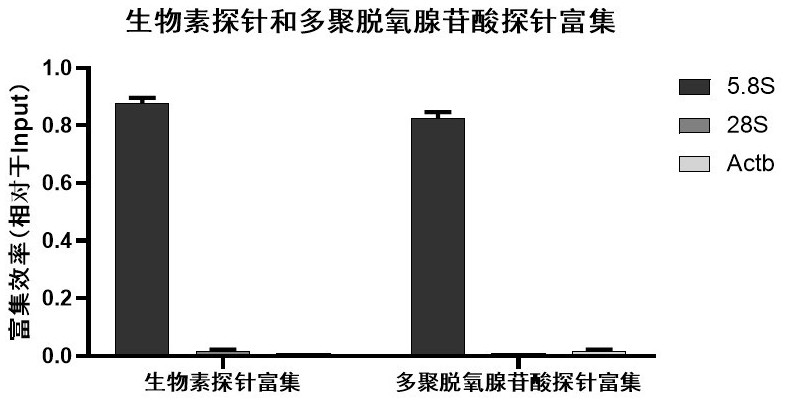
Sequencing method based on gene capture technology
A gene capture and sequencing technology, applied in the biological field, can solve the problems of poor stability of sgRNA and dCas9, difficult sgRNA design, hindering capture efficiency and specificity, etc., and achieve the effects of low synthesis cost, low cost and short time-consuming.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0046] Example 1: Comparison of the application effects of biotin-modified probes and polyA-modified probes in RATE capture technology.
specific Embodiment approach
[0047] In this example, we compared the capture effect of biotin-modified probes and polyA-modified probes on RATE capture technology (see figure 1 with figure 2 ), the target DNA site we picked was double-stranded DNA derived from 5.8S rRNA. The specific implementation is as follows:
[0048] 1) RNA library construction: 1 ug of 293F RNA was constructed using the Hieff NGS® Ultima Dual-modeRNA Library Prep Kit for Illumina (Cat#12252) of Yisheng Biology, and then captured using the RATE-seq process.
[0049] 2) Biotin-modified probe capture
[0050] Table 2
[0051] components Dosage Biotin-5.8S probe (SEQ ID NO: 6) 10-100nM UvsX recombinase 20-200nM UvsY recombinase accessory protein 20-200nM 10× reconstitution buffer 3 µL total capacity 20 µL
[0052] 10× reconstitution buffer including 100-1000 mM tris, 100-3000 mM potassium acetate, 30-500 mM magnesium acetate, 3-30 mM dithiothreitol, 10-200 mM adenosine triphosphate, ...
Embodiment 2
[0068] Example 2: Comparison of the application effects of different 3' end modified probes in the RATE capture technology.
[0069] In this example, we compared the dideoxyribonucleotide modified ddN (SEQ ID NO: 3) and the amino modified NH2C6 (SEQ ID NO: 2) two 3' end modified probes SEQ ID NO: capture at RATE For the technical capture effect, the selected target DNA site is double-stranded DNA derived from 5.8S rRNA. The specific implementation is the same as the biotin-modified probe capture process in Example 1. The result is as Figure 5 As shown, both ddN modification and NH2C6 modification at the 3' end of the probe have good RATE capture efficiency, and the ddN modified probe has better capture efficiency.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com