Novel coronavirus Omicron mutation sequence detection technology based on multiple fluorescent quantitative ARMS-PCR technology and application thereof
A sequence and virus technology, which is applied in the field of detection of the new coronavirus Omicron nucleic acid mutation type, can solve the problems that are not related to the new coronavirus Omicron virus, etc., and achieve the effect of fast and objective detection results, good specificity, and pollution prevention
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0036] Embodiment 1 detection kit and its use
[0037] The kit includes the following components:
[0038] PCR reaction solution, enzyme mixture, mutant gene multiple reaction solution, internal standard reaction solution, positive control 1, negative control;
[0039] PCR reaction solution includes 10× buffer, 25mM MgCl 2 , 10mM dUTP and 10mM dNTPs; the enzyme mixture includes Taq enzyme, reverse transcriptase and UNG enzyme, the ratio of the upstream and downstream primers and probes of the mutant gene multiple reaction solution and the internal standard reaction solution is 4:4:1; The upstream primer is 300 nM, the downstream primer is 300 nM, and the probe is 100 nM.
[0040] See Table 1 for the nucleotide sequences of the primers and probes of the novel coronavirus mutant gene used in this example.
[0041] Table 1 Mutant gene multiplex reaction solution
[0042] 679NK-F TAGGGGCTGAATATGTCAACAACTC SEQ ID No.1 679NK-R CGTGCCCGCCGATGAGACT SEQ ID ...
Embodiment 2
[0099] The sensitivity test of embodiment 2 kit
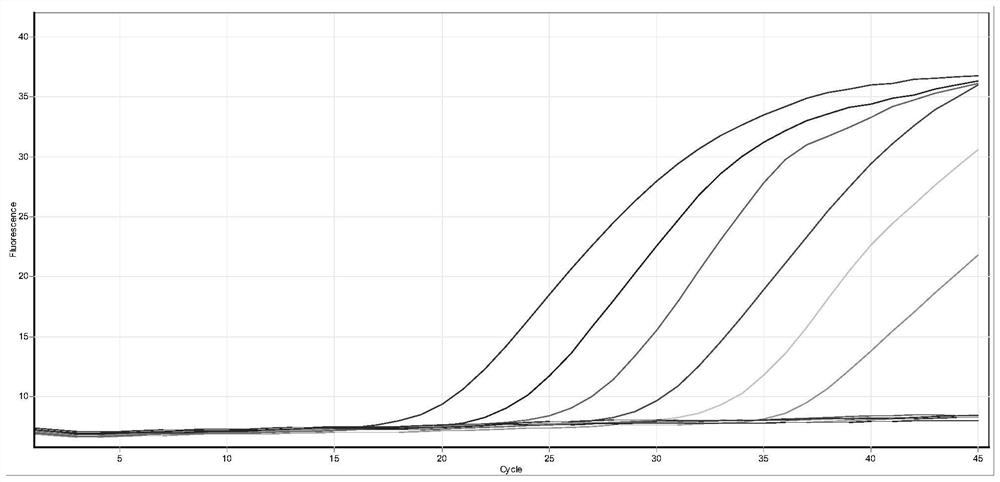
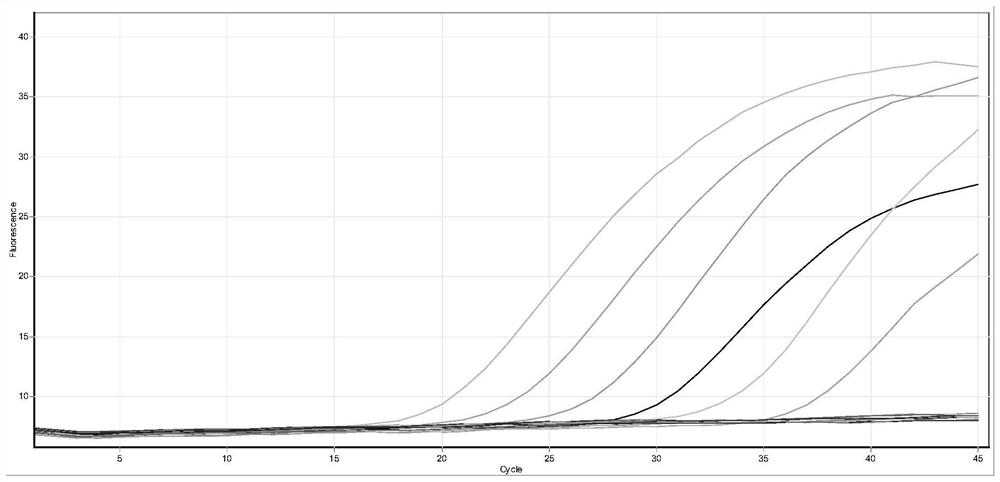
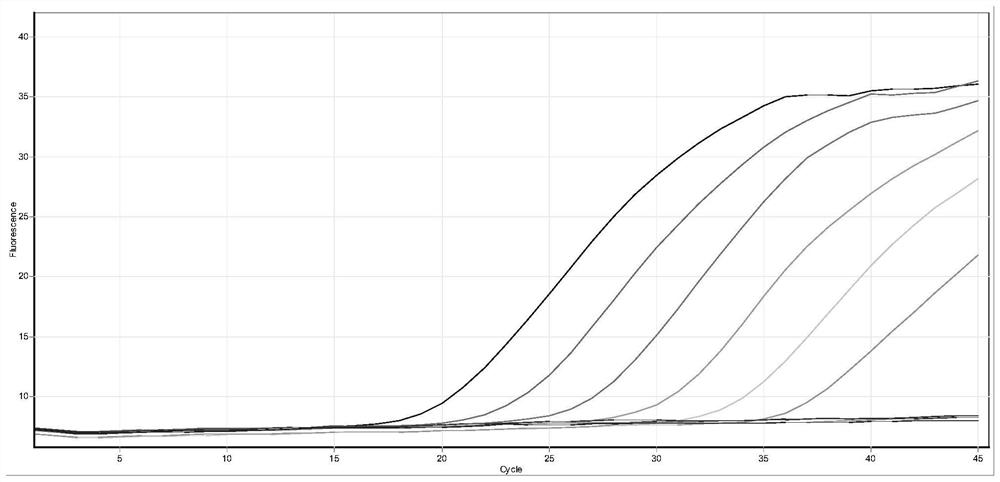
[0100] The positive reference product is a non-replicating lentivirus of the corresponding mutant sequence of the new coronavirus S gene, which is synthesized by a professional organization; 10-fold gradient dilution to 500 copies per milliliter (the amplification curve jumping off on the right in the figure).
[0101] The kit of the present invention is used for detection.
[0102] The test results show that the kit of the present invention has good sensitivity for the detection of the corresponding mutation sequences of the S gene of the new coronavirus (446G>S, 547T>K, 496G>S, 679N>K), reaching 500 copies per milliliter, and CT The value changes in a gradient with decreasing concentration. See figure 1 , figure 2 , image 3 , Figure 4 .
Embodiment 3
[0103] The specificity test of embodiment 3 kits
[0104] In order to detect the specificity of nucleic acid detection kit (fluorescence PCR method) of the present invention, detect rhinovirus, enterovirus, adenovirus, respiratory syncytial virus, influenza virus (A and B type) with it (fluorescence PCR method) , parainfluenza virus, etc.
[0105] The test results show that the detection kit of the present invention can specifically amplify the corresponding mutant sequences (446G>S, 547T>K, 496G>S, 679N>K), without cross-reaction with nucleic acids of other viruses in the respiratory tract, see Figure 5 , Image 6 , Figure 7 , Figure 8 .
[0106] The above results show that the kit of the present invention is generally applicable clinically, has good sensitivity, high specificity, high accuracy rate, and saves time and labor in the operation process.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com