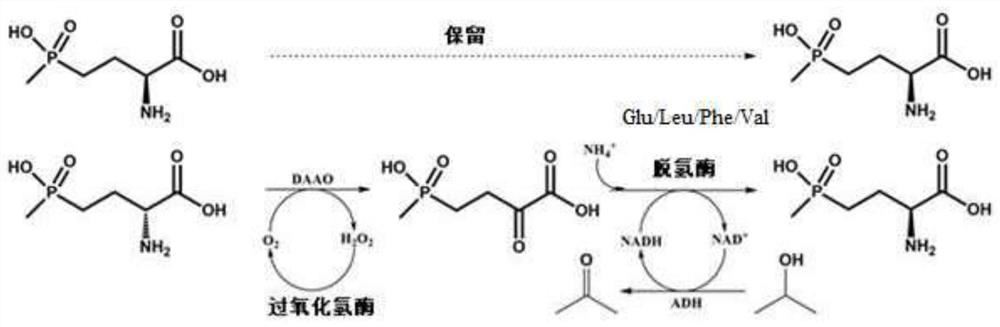
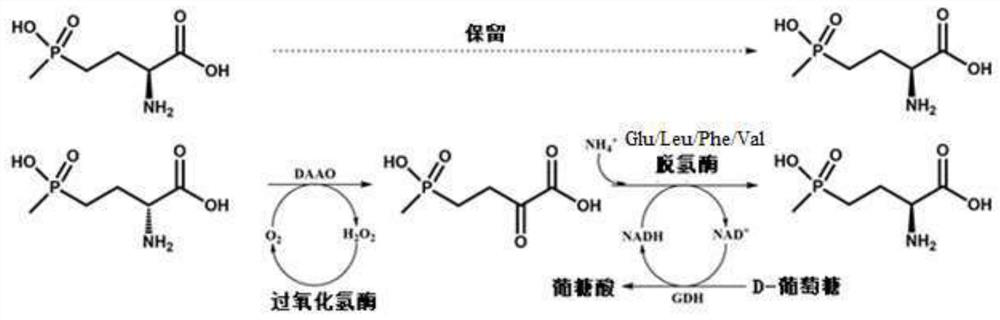
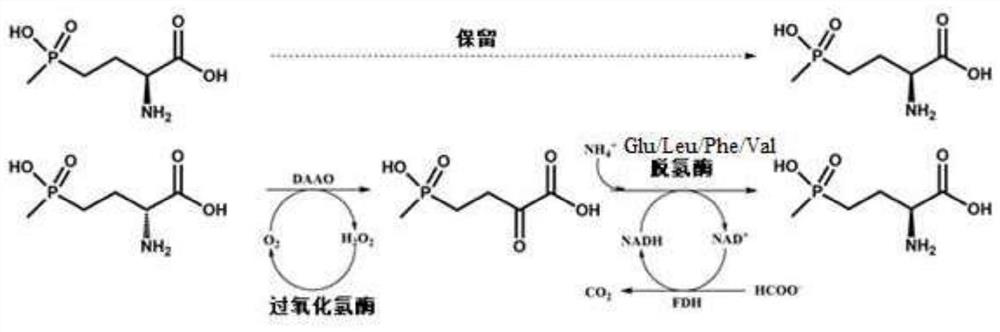
Glu/Leu/Phe/Val dehydrogenase mutant and application thereof in preparation of L-glufosinate
A dehydrogenase and mutant technology, applied in the field of preparation of L-glufosinate-ammonium, can solve the problems of expensive, waste of raw materials, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0089] Embodiment 1: the construction of genetically engineered bacteria
[0090] The gene sequence of D-amino acid oxidase (DAAO, GenBank number: FMSP01000004.1, amino acid sequence shown in SEQ ID NO.1, nucleotide sequence shown in SEQ ID NO.6) derived from Microbotryum intermedium After gene synthesis, the expression plasmid pET-28a(+) was inserted to obtain pET-28a-daao. After sequencing verification, pET-28a-daao was transferred into the expression host E. coli BL21 (DE3) for subsequent expression of the recombinase.
[0091] The sequence of formic acid dehydrogenation (FDH) derived from Lactobacillus buchneri (the amino acid sequence is shown in SEQ ID NO.2, and the nucleotide sequence is shown in SEQ ID NO.7) was inserted into the expression plasmid pET-28a after the whole gene synthesis (+) pET-28a-fdh was obtained. After sequencing verification, pET-28a-fdh was transferred into the expression host E. coli BL21 (DE3) for subsequent expression of the recombinase.
[...
Embodiment 2
[0095] Embodiment 2: the cultivation of engineering bacterium thalline
[0096] The engineered bacteria recombinant Escherichia coli E.coli BL21(DE3) / pET-28a-DAAO, E.coli BL21(DE3) / pET-28a-LAADH, E.coli BL21(DE3) / pET-28a-FDH, E.coli BL21(DE3) / pET-28a-FDH, E. After .coli BL21(DE3) / pET-28a-GDH and E.coli BL21(DE3) / pET-28a-ADH were activated by streaking on the plate, pick a single colony and inoculate it into 10mL LB liquid containing 50μg / mL kanamycin In culture medium, shake culture at 37°C for 10 h. Transfer 2% of the inoculum into 50 mL of LB liquid medium also containing 50 μg / mL kanamycin, culture with shaking at 37 °C until the OD600 reaches about 0.8, add IPTG with a final concentration of 0.1 mM, and shake at 25 °C Cultivate for 12h. After the cultivation, the culture solution was centrifuged at 8000rpm for 10min, the supernatant was discarded, the bacteria were collected, and stored in a -80°C ultra-low temperature refrigerator until use.
Embodiment 3
[0097] Embodiment 3: Construction of D-amino acid oxidase (DAAO) mutant (position 62, position 226)
[0098] On the basis of the wild-type DAAO sequence described in Example 1, the 62nd and / or 226th positions (specifically F62K, M226T) were mutated. The primer sequences of PCR were designed for the mutated mutants at the 62nd and 226th positions of the mutated D-amino acid oxidase sequence, specifically as shown in Table 1:
[0099] Table 1
[0100] serial number Primer name Primer sequence 1 F62KF gattcttgcgggtccaccttggggcaccagttcgctc 2 F62KR gagcgaactggtgccccaaggtggacccgcaagaatc 3 M226TF ggggtctgacgcatcggtagtgcacagcttgac 4 M226TR gtcaagctgtgcactaccgatgcgtcagacccc
[0101] The PCR (25μL) amplification system is as follows:
[0102] Pfu buffer 12.5 μL, primer 2 μL, template plasmid 1 μL, dNTP 0.5 μL, Pfu 1 μL, add ddH 2 O to make up to 25 μL.
[0103] PCR amplification conditions:
[0104] (1) Pre-denaturation at 95°C for ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com